![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
38 Cards in this Set
- Front
- Back
|
Functions of nucleic acids |
Nucleus acids: Storage of genetic info (DNA), transmitted genetic info (mRNA), processing of genetic info (ribozymes), and protein synthesis (tRNA and rRNA) |
|
|
Functions of nucleotides |
Used in monomer form E for metabolism (ATP) Enzyme cofactors (NAD) Signal transduction (cAMP) |
|
|
Nucleotides and nucleosides |
Nucleotides=nitrogenous base, pentose, and PHOSPHATE group Nucelosides= nitrogenous base, pentose C and N atoms on nitrogenous base are numbered in cyclic format |
|
|
Phosphate group |
Negative charge at neutral pH Typically attached to 5’ triphosphates (ATP, GTP, TTP, CTP) 2 of 3 pO used for building nucleic acids form a leaving group |
|
|
Pentose forms differ in some nucleic acids and nucleotides |
Beta-D-ribofuranose in RNA (OH group present) Beta-2-deoxy-d-ribofuranose in DNA (OH group not present) Different puckered conformations of the sugar ring are possible (implications for DNA molecule to fold in space) |
|
|
Nitrogenous bases |
Derivatives of pyrimidine or purine Heteroaromatic, planar Absorb UV light |
|
|
Purines |
Adenine and gaunine Neutral molecules at pH 7 |
|
|
Pyrimidines |
Cytosine, thymine (DNA), Uracil (RNA) Neutral pH at 7 |
|
|
Deoxyribonucleotides |

Know all 4 structures and symbols |
|
|
Ribonucleotides |
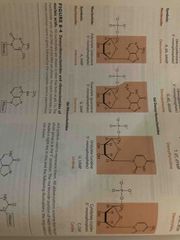
Know structure and symbols |
|
|
Beta-N-glycosidic bond |
Attaches pentose ring to nitrogenous base Formed to the anomeric C in beta configuration Bond is formed to position N1 in pyrimidines and N9 in purines Stable bond, needs to cleaved by acid Relative free rotation around bond: at 0, syn conformation and at 180, anti conformation Anti conformation is found in normal beta DNA |
|
|
Bond rotation possibilities in DNA stretch |

Limited rotation on bond 4 gives rise to ring pucker Can be in endo or exo depending on atoms |
|
|
UV absorption of nucleobases |
UV absorbance is due to pi bonds Excited states decay rapidly due to protection of genetic material |
|
|
Minor nucleosides in DNA |
Modification is done after DNA synthesis (I.e. methylation) -5-methylcytosine: common in eukaryotes and bacteria -N6-methyladenosine: common in bacteria but not in eukaryotes Epigenentic marker: ways to mark own DNA so cells can degrade foreign DNA, way to mark which genes should be active |
|
|
Additional minor nucleosides |
Inosine and pseudouridine |
|
|
Inosine |

Found in the wobble position of the anticodon in tRNA Made by de-aminating adenosine Provides richer genetic code |
|
|
Pseudouridine |

Found in tRNA and rRNA (in arm of tRNA) Common in eukaryotes Made by enzymatic isomerization after RNA synthesis Stabilizes structure of tRNA May help in folding of rRNA |
|
|
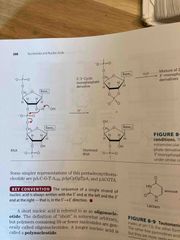
Polynucleotides |
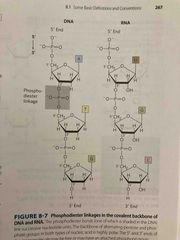
Covalent bonds formed via phosphodiester linkages (negative charge backbone) DNA backbone is stable, hydrolysis by DNAse enzyme RNA backbone is unstable (in water, last a few years; mRNA a few hours in cells) Linear polymers with no branching Directionality: 5’ end is different from 3’ end and read from 5 to 3 |
|
|
Hydrolysis if RNA |
RNA is unstable due to OH- group, over time bonds and cleaves PO3 group (see pic) Hydrolysis is also cleaved by catalytic enzymes (RNase): -rnase p is a ribozyme (enzyme made of RNA) that processes tRNA precursors -dicer is an enzyme that cleaves double stranded RNA into nucleotides Protects from viral genomes |
|
|
Hydrogen binding interactions |

Watson-crick base pairing in DNA A pairs with T C pairs with G Purines pair with pyrimidines Can calculate percentage if you have one: if A is 40%, then t is 40% which means Cand G are both 10 % |
|
|
Watson-crick model of beta DNA |

Antiparallel 10 base pairs per turn Stacking on backbone forming grooves |
|
|
Other forms of DNA |

Beta is most common Alpha is most compact Z is most elongated |
|
|
Complementary of DNA strand |

2 chains differ in sequence and are complementary to each other (read 5-3) They run antiparallel |
|
|
Replication of genetic code |

Strand separation occurs Each strands serves as a template for synthesis of a new strand Catalyzed by enzymes called DNA polymerases A newly made strand has one daughter and one parent strand |
|
|
mRNA: code carrier for proteinds |
Synthesized using DNA template and occurs as single strand -contains ribose instead of deoxyribose -contains uracil instead of thymine One mRNA May code for more than one protein With tRNA, transfers genetic code from dna to proteins |
|
|
Palindrome sequences can form hairpins and cruciforms |

Words or phrases that are the same when read forward or backward |
|
|
RNA molecules have quite complex structures |
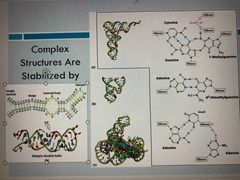
May not be stabilized by Watson-crock base pair interactions PO3 hydrogen binding, bulging |
|
|
DNA denaturation |

Covalent bonds remain intact (genetic code remains intact) Hydrogen bonds are broken (2 strands separate) Base stacking is lost (UV absorbance increase) Can be induced by high temps or change in pH Can be reversible which is called annealing |
|
|
Polymerase chain rxn (PCR) |

Thermal DNA densturating: exist as double helix but high temps unwinds, low temp re-anneal This is basis for PCR
|
|
|
Factors affecting DNA denaturation |
Tm is the midpoint to get to denaturation Higher GC increases Tm (base composition) Longer DNA has higher Tm (DNA length) High salt increases Tm (pH change |
|
|
Deamination |

Slow reactions Large number of residues Significant net effect: 100 C -> U events daily |
|
|
Depurination (mutation) |

Glycosidic bond is hydrolysized Free residue floating around Ribose could then open up and become linear Happens 10,000 times a day |
|
|
Oxidative and chemical mutagenesis |
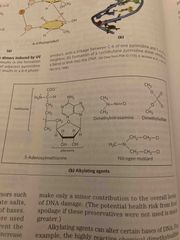
Oxidative damage: hydroxylation of gaunine, mitochondrial DNA is most susceptible Chemical alkylation: methylation of gaunine Cells have mechanisms to correct most of these modifications |
|
|
Radiation-induced mutagenesis |
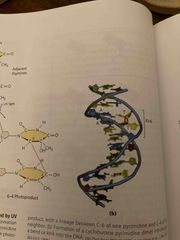
UV light induced dimerization of pyrimidines; may be skin cancer cause Ionizing radiation: X-rays cause ring opening and strand breaking (difficult to repair) Cells can repair some modifications but others may cause mutations Accumulation of mutations is aging and cancer |
|
|
E source of nucleotides |

Evolutionary chose ATP for us but could be any of pics |
|
|
Coenzymes and nucleotides |

Coenzyme A |
|
|
Regulatory molecules nucleotides |

Signal to cells |
|
|
H |
H |

