![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
37 Cards in this Set
- Front
- Back
|
Enzymes |
Catalysts that increase reaction rates without being used up Most are globular proteins (some RNA also catalyze) Studies of enzyme is oldest field |
|
|
Holoenzyme v apoenzyme |
Enzyme require molecules called cofactors or coenzymes Forms complex Vs no cofactors
|
|
|
Why biocatalyst I’ve inorganic catalyst |
Greater reaction specificity and avoids side products Milder reaction conditions, conducive to cell conditions Higher reaction rates Capacity for regulation (biological pathways)
|
|
|
Metabolites have many potential pathways of decomposition |
Enzymes make the desired one most favorable |
|
|
Classes of enzymes |
TOHILL Transferases: group transfer reactions Oxidoreductases: transfer of e- Hydrolases: hydrolysis reactions Isomerases:transfer of groups in molecules to yield isomeric forms Lyases: cleavage of C-C, C-O, C-N bonds, leaving double bonds or rings, or addition of groups Ligases: formation of C-C, C-S,C-O, C-N bonds by condensation, coupled to cleave ATP or similar cofactor |
|
|
Enzyme do not affect |
Equilibrium or free energy of reaction Help overcome activation barriers Increase reaction rates by decreasing free energy |
|
|
How to lower free energy |
Uncaralyzed bimolecular reactions: 2 free reactants —> single restricted transition state (entropically unfavorable) Uncatalyzed unimolecular reactions: flexible reaction —> rigid transition state conversion (entropically unfavorable) Catalyze reaction: enzyme uses binding energy of substrates to organize reactants to fairly rigid complex, entropy cost is paid in binding, rigid reaction is entropically neutral |
|
|
Free E |
See pic |
|
|
Diagram |
Pic |
|
|
Enzymes bind transition states best |
Enzymes active sites are complimentary to the transition state of the reaction Bind transition states better than substrates Stronger transition states lower the activation E barrier |
|
|
Catalytic mechanisms |
Acid-base: give and take protons Covalent: change reaction paths Metal ion: use redox cofactors, pKa shifters |
|
|
Enzyme Kinetics |
Study of the rate at which compounds react Enzymatic Rate is affected by: enzyme, substrate, effectors, temp |
|
|
Why study enzyme kinetics? |
Quantitative description Determine order of binding substrates Elucidate acid-base catalyst Understand catalytic mechanism Understand reg of activity |
|
|
Kinetic equations |
Start w model mechanism Identify constraints and assumptions Algebra |
|
|
Simplest model mechanism |

One reactant, one product, no inhibitors |
|
|
Id constraints and assumptions |
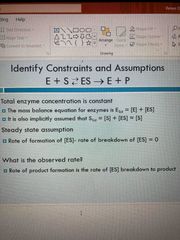
Total enzyme concentration is constant |
|
|
Algebra: the Michaelis menten equation |
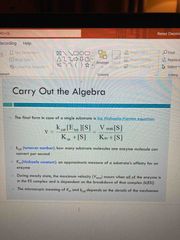
Kcat: how many substrate molecules one enzyme molecule can convert per second Km: (Michaelis constant) approx. measure of a substrates affinity for an enzyme |
|
|
Equations for exam |
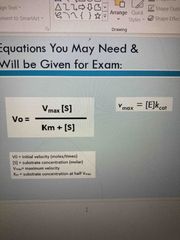
A |
|
|
How to do kinetic measurements |
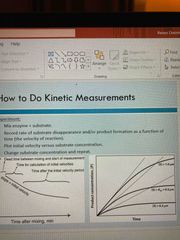
Mix enzyme and substrate Record rate of substrate disappearance and/or product formation of time Plot initial velocity vs substrate concentration Change substrate concentration and repeat |
|
|
Determination of kinetic parameters |
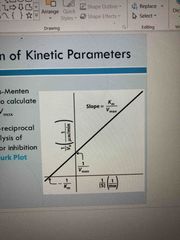
A nonlinear plot used to calculate Km and Vmax A linear double/reciprocal plot is good for analysis of 2 substrate data or inhibition (line weaver plot) |
|
|
Step 2 |
Ser and his generate ion that attacks peptide carbonyl group Forming short lived negative intermediate |
|
|
Sequential |
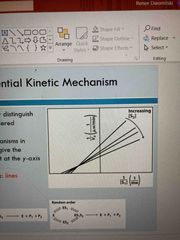
Cannot easily distinguish random from ordered Will give intersection at y axis |
|
|
Ping pong |
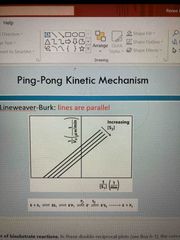
Lines are parallel |
|
|
Step 5 |
Collapse of intermediate and second product formed: a carboxylate anion and displaces ser |
|
|
Competitive inhibition |

Competes with substrate for binding Binds to active site Does not affect catalysis No change in Vmax, but increase in Km Lone intersect on lineweaver-burk |
|
|
Uncompetitive inhibition |

Only binds to ES complex Doesn’t affect substrate binding Inhibits catalytic function Decrease in Vmax and Km Lines are parallel |
|
|
Mixed inhibition |

Binds enzyme with or without substrate Binds to regulatory site Inhibits both substrate binding and catalysis Decrease in Vmax, change in Km Lines intersect |
|
|
Chymotrypsin |
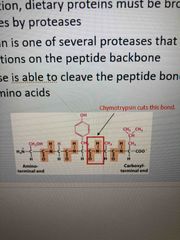
During digestion, proteins broken down into small peptides by protease This is one of them Able to cleave peptide bond adjacent to aromatic AAs |
|
|
Step 1 |
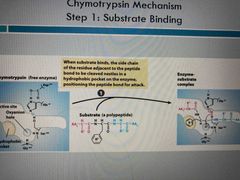
Substrate binding Side chain in hydrophobic pocket |
|
|
Step 2 |

Ser and his generate ion that attacks peptide carbonyl group Forming short lived negative intermediate |
|
|
Step 3 |
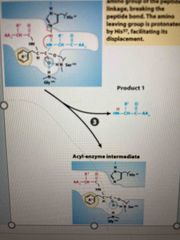
Collapse of intermediate Reformation of double bind with C breaking peptide bond Amino leaving group is protonated |
|
|
Step 4 |
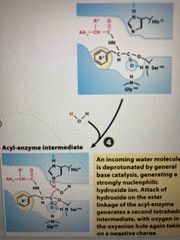
Water molecule is deprotinated forming OH ion OH ion breaks water link forming second intermediate O is again negative in the oxyanion hole |
|
|
Step 5 |
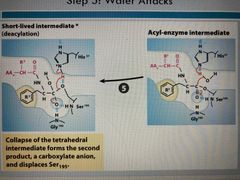
Collapse of intermediate and second product formed: a carboxylate anion and displaces ser |
|
|
Step 6 |
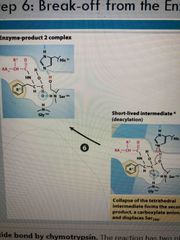
Break off enzyme |
|
|
Step 7 |
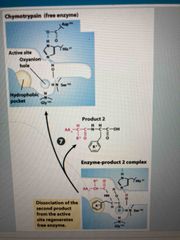
Dissociation of second product regenerates free enzyme |
|
|
Sequential |
Cannot easily distinguish random from ordered Will give intersection at y axis |
|
|
Proteases cleaving |
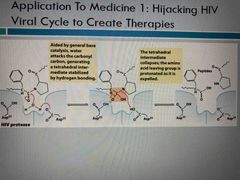
Back (Definition) |

