![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
52 Cards in this Set
- Front
- Back
|
In the case of a viral infection, what neutralizes the extracellular virus particles? What produces this?
|
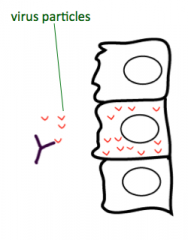
Antibodies (made by B cells)
|
|
|
In the case of a viral infection, what eliminates the source of the virus?
|
T cell response
(Humoral B cell response cannot do this) |
|
|
Where are viral proteins made?
|
Within the host cell, by the host's own protein synthesis machinery
|
|
|
Why must an effective immune system test both vacuolar and cytoplasmic compartments?
|
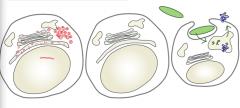
- Viral fragments (red) are in cytoplasm
- Bacterial fragments (green) are in vacuolar compartments |
|
|
Through what process do viral and bacterial proteins get degraded?
|
Routine protein turnover - all proteins in a cell (both cellular and foreign) are degraded to fragments/peptides via this process
|
|
|
What happens to the peptide fragments of viral and bacterial origin after being degraded by routine protein turnover?
|
Peptide fragments are presented to T cells on Class I (viral) or Class II (bacterial) MHC molecules = Antigen Presentation
|
|
|
What is the inside of a vacuole equivalent to?
|
Outside of the cell
|
|
|
What is the primary difference between Class I MHC and Class II MHC molecules?
|
- Class I - sample inside of a cell (cytoplasm)
- Class II - sample outside of a cell/vacuoles |
|
|
What is the process of a molecule being presented on a Class I MHC antigen?
|
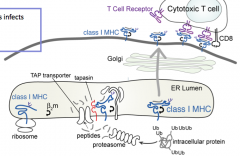
1. Proteins must be tagged for destruction
2. Proteolysis must occur to generate peptides of appropriate size 3. Peptides must be delivered to class I MHC molecules 4. Peptides must bind to class I MHC molecules 5. Peptides must be displayed to T cells in context of class I MHC molecules |
|
|
How are proteins tagged for proteolysis?
|
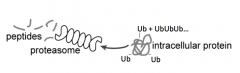
- Ubiqutin (small 8 kDa protein) added to protein destined for degradation (on Lysine residues)
- Forms a Ub chain (polyubiquitination) |
|
|
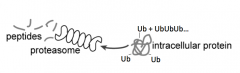
How are peptides generated?
|
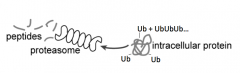
- Protein that is labeled w/ Ubiquitin is recognized by Proteasome
- Proteasome cuts up protein to release peptides |
|
|
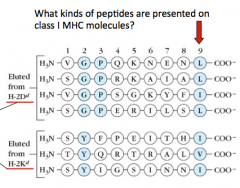
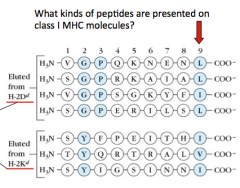
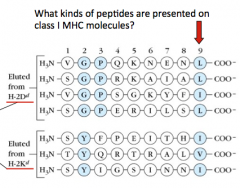
What are some characteristics of the peptides that are presented on Class I MHC molecules?
|
- 9 peptides long
- End w/ a hydrophobic residue (L, I, V, F) |
|
|
If a cell is infected with a virus, how does it enrich for peptides that are suitable for loading onto class I MHC molecules?
|
- Proteasomes have 3 types of protease activity that are suitable for producing peptides that are presented on class I MHC molecules
- Chymotrypsin-like proteases, Trypsin-like proteases, and Caspase-like proteases - Chymotrypsin-like proteases cleave proteins so that they end w/ a hydrophobic residue (L, F, I, V) |
|
|
Which type of proteases cleave proteins to generate peptides that end with hydrophobic residues? What is the significance of this?
|
Chymotrypsin-like proteases (end w/ L, F, I, or V) - these peptides are suitable for binding to Class I MHC molecules
|
|
|
What must happen to a protein before it can be processed by the proteasome? Why?
|
- Ubiquitin must be removed by isopeptidases
- Protein must be unfolded by unfoldases - The diameter of the center of the proteasome is only 13A (so can't fit a folded protein w/ ubiqutin on it) |
|
|
What size of peptides does the proteasome generate?
|
4-20 AA residues
|
|
|
What happens to peptides after they are cleaved by the proteasome?
|
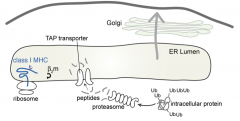
TAP: Transporter associated w/ Antigen Processing transports peptides into the ER
|
|
What is the preferred substrate of the TAP (Transporter associated w/ Antigen Processing)?
|
- Only peptides
- Favors peptides ending w/ L, I, V, or M (hydrophobic) - 6-15 AA residues |
|
|
What are the molecular sieves that limit the size of peptides that are presented onto Class I MHC molecules?
|
- Proteolysis by proteasomes generates peptides 4-20 residues
- TAP transporter selects for subset ending w/ L, I, V, or M that are 6-15 residues long - Binding to class I MHC molecule has strict size restrictions of 8-10 residues (image shows that many are 9 AA long) |
|
|
Which proteins help keep the TAP transporter in close proximity to the Class I MHC molecule? Why?
|
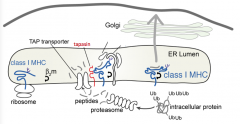
- Tapasin and light chain β2m
- Makes sure that Class I MHC molecules are close to the incoming peptides |
|
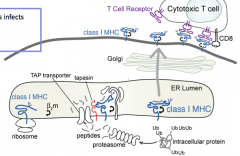
When does this process happen?
|
At all times, regardless of whether foreign proteins are present
|
|
|
What happens when a virus infects a cell?
|
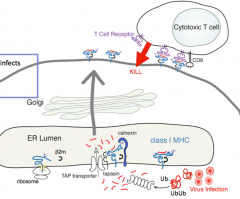
1. Protein tagged for destruction (ubiquitin)
2. Proteolysis (peptides ending in L, I, V, or M) 3. Delivery of peptide (selection for L, I, V, or M enders) 4. Binding of peptide (chaperone-mediated) 5. Transport to cell surface and presentation to T cells (which proceed to kill the cell presenting the peptide) |
|
|
What is the structure of the Class I MHC molecule?
|
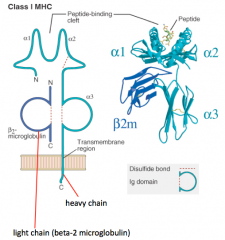
- Heavy chain makes peptide-binding cleft on its own (α1, α2, and α3)
- Light chain (β2-microglobulin) is much smaller - Peptide is critical to structure (without it molecule falls apart) |
|
|
In which kind of MHC molecule does a single heavy chain form the entire peptide-binding cleft?
|
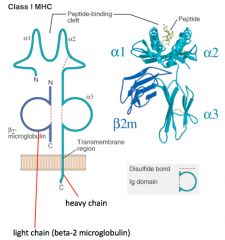
Class I MHC
|
|
|
What is the structure of the Class II MHC molecule?
|
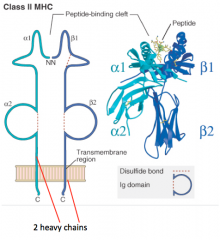
- 2 Heavy chains contribute equally to peptide-binding cleft (α1/α2 and β1/β2)
- No light chains - Peptide is critical to structure (without it molecule falls apart) |
|
|
In which kind of MHC molecule do two heavy chains form the peptide-binding cleft?
|
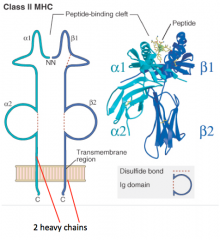
Class II MHC
|
|
|
What happens if there is no peptide bound to a MHC molecule?
|
MHC molecules fall apart, therefore the peptide is considered a subunit of the MHC molecule
|
|
|
What secondary structures form the peptide binding cleft of MHC Class I molecules?
|
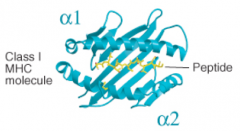
α1 and α2 both contribute α helices and β pleated sheets (both from heavy chain)
|
|
|
What secondary structures form the peptide binding cleft of MHC Class II molecules?
|
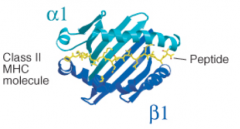
- α1 and β1 each contribute α helices and β pleated sheets
- α1 and β1 are from separate heavy chains |
|
|
Class I MHC molecules hold what length of peptides in their binding cleft?
|
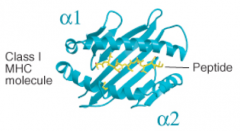
8-10 residues
|
|
|
Class II MHC molecules hold what length of peptides in their binding cleft?
|
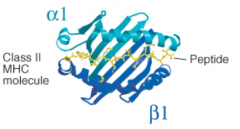
10-16 residues, but can be longer, up to 30+
|
|
|
How does tagging of proteins differ for Class I vs Class II MHC molecules?
|
- Class I - proteins must be tagged (ubiquitination)
- Class II - no tagging of proteins |
|
|
How does delivery of peptides differ for Class I vs Class II MHC molecules?
|
- Class I - Peptides must be delivered to MHC molecules
- Class II - No topological barriers, delivery not an issue |
|
|
How does the location of binding of peptides to MHC Class I vs Class II molecules differ?
|
- Class I - in ER
- Class II - in endocytic compartment (not ER) |
|
|
Class I vs Class II MHC molecules bind to which T cells?
|
- Class I - CD8 T cells
- Class II - CD4 T cells |
|
|
In the Class II MHC molecule pathway, how do you know what is going to be lysed?
|
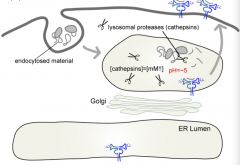
Anything that is delivered to the lysosome - does not have to be tagged
|
|
|
How are antigens lysed into peptides for the Class II pathway?
|
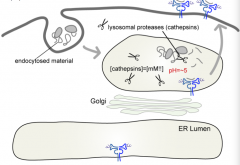
Lysosomal Proteases (Cathepsins - cystein proteases)
|
|
|
What is the concentration of Cathepsin / Lysosomal proteases in the lysosome?
|
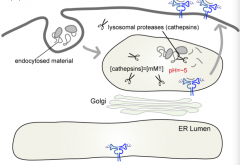
On the order of mM (this is very concentrated)
|
|
|
What is the pH in a lysosome?
|
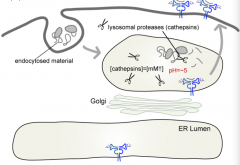
pH = ~5
|
|
|
How do Class II MHC molecules get into the lysosome (where they load peptides)?
|
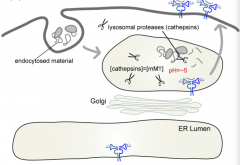
- Start at ER
- Pass through Golgi - Lysosome (where they load peptides) - Plasma membrane |
|
|
How do you prevent peptides in ER destined for Class I molecules from binding to class II molecules?
|
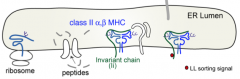
Invariant chain (Ii) sits in peptide groove and blocks binding of peptides in ER
|
|
|
What is Invariant chain (Ii)?
|
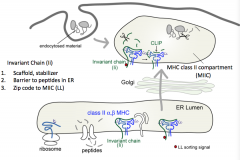
- Scaffold, stabilizer
- Barrier to peptides in ER (sits in peptide groove) - Contains zip code to MIIC (MHC Class II Compartment) |
|
|
What part of the Invariant chain (Ii) peptide sends the MHC Class II molecules to the lysosome? What is the name of this compartment?
|
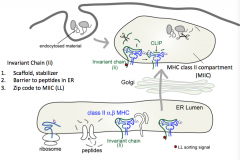
- LL sorting signal (two lysine residues)
- Sends MHC Class II to MHC Class II compartment (MIIC) |
|
|
What is the structure of Invariant Chain (Ii)?
|
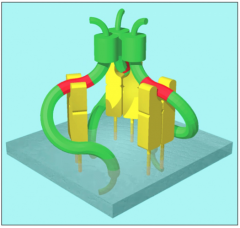
Trimer w/ 3 class II molecules and 3 invariant chains
|
|
|
How do you remove the Invariant Chain (Ii) from the MHC class II groove?
|
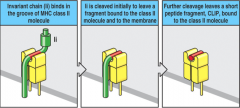
- Cathepsin initially cleaves Ii to leave a fragment bound to the molecule and the mebrane
- Further cleavage leaves peptide fragment, CLIP in groove - HLA-DM catalyzes exchange of CLIP and antigenic peptide |
|
|
What is the function of HLA-DM?
|
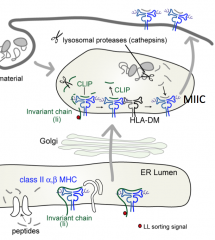
Catalyzes exchange of CLIP (peptide remaining in Class II MHC groove) for an antigenic peptide
|
|
|
Is the groove of a Class II MHC molecule usually empty or full?
|
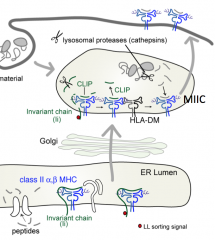
Usually full - only empty when HLA-DM catalyzes the removal of CLIP (before binding of antigenic peptide)
|
|
|
Why must a peptide almost always be found in the groove of the Class II MHC molecule?
|
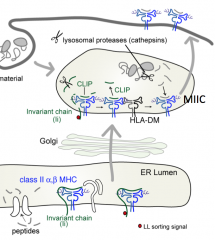
Without the peptide in the groove the Class II molecule is destabilized (black), it unfolds, and is destroyed by proteases in the lysosome
|
|
|
Can self-peptides be presented by Class II MHC molecules?
|
Yes
|
|
|
What are the specialized antigen presenting cells for Class II MHC molecules?
|
- B cells
- Dendritic cells - Macrophages |
|
|
On what kind of cells are Class I MHC molecules expressed?
|
All nucleated cells
|
|
|
On what kind of cells are Class II MHC molecules expressed?
|
Only on specialized antigen presenting cells (B cells, Dendritic cells, and Macrophages)
|

