![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
29 Cards in this Set
- Front
- Back
|
What is Proteolysis? |
Hydrolysis of dietary proteins, recycling of AA, activation of pro-enzymes and hormones. Used up in cell migration, metalloproteases and ECM. Degradation of regulatory proteins, damaged proteins, misslocalized proteins, missfolded proteins, foreign proteins, proteins in stoichiometric excess Non-Specific Proteolysis = digestion of dietary protein, extracellular Specific = degradation of specific proteins, intracellular |
|
|
What is non-specific proteolysis? |
-non specific dietary protein degradation-digestion of proteins-> AA-Pepsin (stomach); Elastase, Trypsin, Chymotrypsin, Carboxypeptidase (pancreas); enteropeptidase, aminopeptidase (Sm Int)-extracellular, in the lumen -eat ~100g, rest controlled degradation -excess burned as fuel (into CO2) |
|
|
Controlled/ specific proteolysis? |
intracellular protein degradation (~400g/day), adds to protein synthesis needs (porphyrin, creatin, neuortansmitters, purines, pyrimidines, other nitrogen containing compounds etc) |
|
|
how is proteolysis controlled? |
-proenzymes -compartmentalization |
|
|
what is protein 1/2 life? |
- average life of a protein -physical (damaged) or functional (obsolete/ no longer necessary) -protein turnover |
|
|
how does protein folding occur? |
-down entropically favorable cascade -uses chaperones and chaperonines to assist -direction toward native fold is thermodynamically most favorable -dissolution increases entropy of water, favorable |
|
|
What are chaperones? |
-HSP 70 : binds to nacent polypeptide (dusing protein synthesis), prevents it from folding back onto itself, assists in correct folding. Unbinds at cost of ATP. Can bind to undo aggregation or deliver to Chaperonine if still miss folded -HSP 40: also unbinds art cost of ATP, assists in correct folding. Both prevent aggregation of missfolded proteins and the miss folding of nacent protein act on folding and miss folded proteins |
|
|
What are chaperonines? |
-larger three unit structures that help to fold missfolded proteins (at cost of ATP)delivered by chaperones, unbind at the cost of ATP or begin proteolysis if still missfolded |
|
|
three main routes to get rid of a miss folded protein |
-tagging, and dielivery to autophagosome which then fuses with a lysosome -aggregation, partial unfolding followed by a chaperone ->chaperone mediated autophagy -ubiquitination and degredation by a proteosome |
|
|
some post translational modifications that are possible to do on a protein |
-add: iron, sugar, disulfide bond formation(oxidation), phosphorylation, ubiquitination |
|
|
overview of Ubiquitin-Proteosome system |
-attach Ub to lysine of protein (E1 uses hydrolysis of ATP->AMP +PPi to bind Ub, passes to E2 which binds E3) -send ubiquitinated protein to Proteosome -proteolysis is energetically costly, recycles AA and Ub |
|
|
how are proteins degraded in a specific/controlled manner |
-conditional expression of targeting factors, a regulatory protein (e.g. p53, Hif1, NFkB, IkBå, topoisomerase etc) -modification of substrate (Ub tag) |
|
|
describe Ubiquitin |
-active Glycine COO- end -7 lysine residues -small hydrophobic patch -most conserved protein in evolution -always expressed as a 'fusion protein'(Ub's in tandem or fused to specific ribosomal protein) and then peptide bonds are hydrolyzed into monomers by Ub carboxy terminal hydrolase -76 AA, 8.5kDaltons -binds its terminal Glycine's COO- to NH of LYSINE |
|
|
describe Ubiquitination |
-PolyUb chain is hydrolyzed (UCTH) -E1 (Ub Activating enzyme) binds Ub Monomer at cost of ATP-> AMP +PPi -E1 passed Ub to E2 (Ub Conjugating Enzyme) -E2 and E3 (Ub-Protein Ligase) dimerize and substrate is tagged * -Mono/Di Ub of membrane proteins for endocytosis or cell trafficking (the balance between E2E3 : DeUbProtein (DUB) determines if/how long cycle repeats) -polyUb for cytosolic proteins to be sent to Proteasome for degradation -at cost of ATP Ub is recycled |
|
|
describe E1 |
-Ub Activating Enzyme -Binds 2 Ub (at cost of ATP) as a Ub-Adenylate via a thioester bond |
|
|
describe E2 |
-Ub Conjugating Enzyme -mediates transfer of Ub from E1 to either E3 or Substrate -binds Ub via a thioester bond (catalytic Cystein: COOS) |
|
|
describe E3 |
-Ub-Protein Ligase, recognizes protein substrate and E2 -mediates transfer of Ub to Substrate from E2's Cystein theioester (Ring) or takes Ub from E2 and then passes it to Substrate (Hect) via an isopeptide bond (Gly-Lys: COON) |
|
|
what is the significance of a polyUb chain? |
-distinct Lys on Ub can support being Ubiquitinated, -designates cytosolic proteins to Proteosome for degradation -creates hydrophobic stripe that is recognized by proteosome (all the distinct hydrophobic patches come together), primary determinant for interacting with proteosome -7∆ Lysines, therefore at least 7∆ types of signals are possile (Ub on K6 or K63= DNA repaire, K11 or K48 = degredation, K33= stress response etc) |
|
|
what is combinatorial diversity? |
the different combination of different types of of E2 (~12 kinds) and E3 complexes (~1000) that allows the binding of ~1million different proteins in cell |
|
|
describe a eukaryotic proteosome |
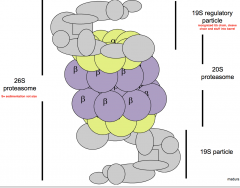
-two regulatory 19S ends that bind hydrophobic stripe of PolyUb and releases them, contain unfolding enzymes (ATP dependent) -central 20S catalytic core, three hydrolytic activities sequestered within, gated entry, four stacked seven subunit rings form a barrel -without the proteasome the cell will die (since cant get enough AA) |
|
|
what are some energy dependent and energy independent steps of the Ub-Proteosome system? |
-Energy Dependent: Ub activation, proteosome assembly, chaperone mediated refolding, substrate unfolding -Energy Independent: Thioester cascade (E1-E2/E3), hydrolysis of of proteosome, de-Ub protein, Ub processing |
|
|
what causes the up/down regulation of proteasome? |
Nutrient Starvation and Rapamycin inhibit TORC1(master growth controller of metabolic processes) causing upregulation of proteasome subunit and levels, increaseing protein degradation and [AA] |
|
|
how are membrane proteins recycled: |
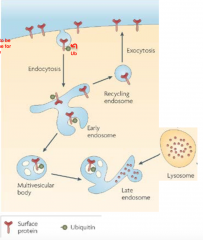
-monoUb of protein -> endosomal or lysosomal pathway -lysosomes, membrane bound organelles contain acid hydrolase for digestion of protein |
|
|
what are two key components in ER quality control? |
ERAD- Endoplasmic Reticulum Associated protein Degradation (dislocon to get missfolding glycoprotein out of ER to proteasome) UPR- Unfolded protein regulation(when ERAD fails and missfolded proteins accumulate, stops translation and/or induces apoptosis) |
|
|
describe the ERAD pathway |
-Recognition: chaperones in ER recognize missfolded protein, help refold if possible and send to Golgi (note: once a missfolded protein is in Golgi or beyond must go to lysosome) -Retention: terminally missfolded glycoproteins recognized by chaperone(BiP?),, sent back out through Dislocon into cytoplasm. De-glycosylated -Ubquitiated and sent to proteasome or selective autophagy |
|
|
describe UPR |
Unfolded protein response is a signaling pathway protein folding status in ER. When misfolded proteins acumulte in the ER, UPR sends a signal into the cytosol and nucleus for transcriptional induction of UPR genes, attenuation of global protein synthesis and ERAD pathway. If ERAD still fails, then signals to induce Apoptosis |
|
|
what are some Ub linked diseases? |
Tumorogenesis, cancer HPV Parkinsons Inflammatory response Alzheimers Prions Lysosomal storage diseases like Gaucher Angelman's syndrome |
|
|
describe tumorgenesis |
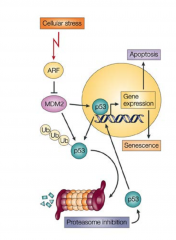
|
|
|
describe HPV hijacking system |
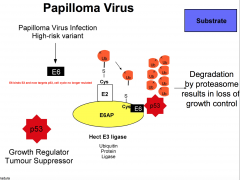
|

