![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
73 Cards in this Set
- Front
- Back
|
Asexual Reproduction |

|
|
|
Sexual Reproduction |

|
|
|
Natural Selection Flow Chart |
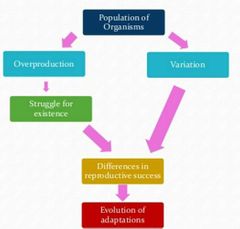
|
|
|
Forces of Evolutionary Change |
Natural Selection
&
Genetic Drift |
|
|
Natural Selection |

|
|
|
Genetic Drift |

|
|
|
Mutation |
Heritable change in DNA sequence that can lead to a change in phenotype (observable properties of an organism). |
|
|
Wild-Type Strain |
Typically refers to strain isolated from nature. “Wild-type” can also refer to just one gene. |
|
|
Mutant |
A cell or virus derived from wild type that carries a nucleotide sequence (genotype) change. Observable properties (phenotype) may also be altered. Can be obtained from parental strain derived fromwild-type. |
|
|
Genotype |
Genotype is designated by italicized (three lowercase letters + capital letter) for a gene (e.g., hisC). |
|
|
Mutations |
Mutations are designated with numbers referring to order of isolation (e.g., hisC1, hisC2). |
|
|
Phenotype |
Phenotype is designated by capital letter + two lowercase letters and +/– to indicate presence/absence (e.g., His+ or His–). |
|
|
Spontaneous Mutations |
Those that occur without external intervention. Most result from occasional errors by DNA polymerase during replication. |
|
|
Induced Mutations |
Those made environmentally and deliberately. Can result from exposure to natural radiation or chemicals that chemically modify DNA. |
|
|
Point Mutations |
Mutations that change only one base pair. Can lead to single amino acid change in a protein, an incomplete protein, or no change at all. |
|
|
Types of Base-Pair Substitutions |
• Missense
• Nonsense
• Silent Mutations
Not all mutations change polypeptides due to degeneracy. |
|
|
Silent Mutations |
Do not affect sequence of encoded polypeptide or phenotype.
Almost always third base of codon. |
|
|
Missense Mutation |
Changes sequence of amino acids in polypeptide (e.g., UAC to AAC).
If at a critical location, especially active site, could alter activity.
Not all missense mutations lead to dysfunction. |
|
|
Nonsense Mutation |
Leads to stop codon. Typically results in truncated (incomplete) protein that lacks normal activity. |
|
|
Base-Pair Substitutions Visual |
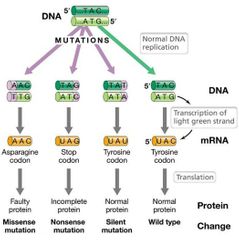
|
|
|
Transitions |
Purines (A/G) substituted for other purines. |
|
|
Transversions |
Purines substituted for pyrimidines (C/T) or vice versa. |
|
|
Frameshift Mutations |
Deletions or insertions that result in a shift in the reading frame. Insertion/deletion of three base pairs adds/deletes an amino acid, which usually is not as bad. Scrambles entire polypeptide sequence downstream. |
|
|
Insertions/Deletions... |
... can result in gain/loss of hundreds to thousands of base pairs. Often result in complete loss of gene function. May arise from errors during genetic recombination. Large insertions may be due to transposable elements. |
|
|
Frameshift Visual |
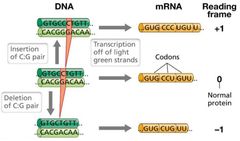
|
|
|
Mutation Rates Pt. 1 |
Mutation rates depend on frequency of DNA changes and efficiency of DNA repair.
For most microorganisms, errors in DNA replication occur at a frequency of 10^-6 to10^-7 per kb. Since a typical gene is ~1 kb, frequency of mutation in a single gene is in the same range. |
|
|
Mutation Rates Pt. 2 |
Eukaryotes have 10-fold lower error rates.
DNA viruses have error rates 100–1000x greater.
RNA viruses even higher due to less proofreading and lack of RNA repair mechanisms. |
|
|
Mutation Rates Pt. 3 |
Single base errors more likely to lead to missense than nonsense mutations because most substitutions encode amino acids. Next most frequent single base change leads to silent mutation. Can increase pool of mutations by treatment with mutagenic agents or under high stress. |
|
|
Mutagens |
Chemical, Physical, or Biological agents that increase mutation rates. |
|
|
Chemical Mutagens and Radiation Pt. 1 |
Nucleotide Base Analogs:
Resemble nucleotides but have faulty base-pairing.
Replication errors occur at higher frequencies due to incorrect base pairing. |
|
|
Chemical Mutagens and Radiation Pt. 2 |
Induce chemical modifications.
Example: Alkylating agents such as nitrosoguanidine introduce changes in replicating or nonreplicating DNA. |
|
|
Chemical Mutagens and Radiation Pt. 3 |
Chemical mutagens that cause frameshift mutations.
Example: Intercalating agents such as acridines push two base pairs apart, triggering insertions or deletions. |
|
|
Chemical Mutagens and Radiation Pt. 4 Nonionizing Radiation |
(e.g., Ultraviolet [UV])
Purines and pyrimidines strongly absorb UV.
Pyrimidine dimer (two adjacent Cs or Ts on the same strand become covalently bonded) is primary effect of UV radiation.
Killing cells by UV is primarily due to effect on DNA. |
|
|
Chemical Mutagens and Radiation Pt. 5 Ionizing Radiation |
(e.g., X-rays, cosmic rays, and gamma rays) More powerful than UV. Ionize water, forming free radicals such as hydroxyl radical (OH·) that damage macromolecules, leading to double- or single-stranded breaks and rearrangements or large deletions. |
|
|
Electromagnetic Spectrum Visual |
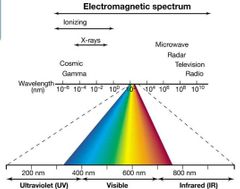
|
|
|
Homologous Recombination |
(Their effect of on genotype) In bacteria, genetic recombination occurs after transformation, transduction, or conjugation. To detect exchange of DNA, recombinant cells must phenotypically differ from both parents. Usually use recipient strains lacking a selectable characteristic that recombinants gain. |
|
|
Transformation |
Genetic transfer process by which free DNA is incorporated into a recipient cell and brings about genetic change. |
|
|
Competence in Transformation |
Competent: a cell that can take up DNA and be transformed; genetically determined. |
|
|
In Naturally Transformable Bacteria... |
... competence is regulated, and competence-specific proteins uptake and process DNA. Quorum sensing in Bacillus subtilis. Quorum sensing, chitin sensing, catabolite repression in Vibrio cholerae. High-efficiency natural transformation is rare in Bacteria. |
|
|
In Other Strains... |
... specific procedures are necessary to make cells competent (e.g., treatment of E. coli with high Ca+2 and chilling). Electricity can be used to force cells to take up DNA (electroporation). |
|
|
Uptake and Integration of DNA in Transformation Pt. 1 |
Natural transformation starts with reversible DNA binding that becomes irreversible.
Competent cells bind up to 1000x more DNA than noncompetent cells. Linear DNA first bound by DNA-binding protein similar to pilus. |
|
|
Uptake and Integration of DNA in Transformation Pt. 2 |
Either ds fragment taken in or nuclease degrades one strand and other is taken in. Competence-specific protein binds and protects DNA. RecA integrates DNA. In contrast, plasmid DNA must remain ds and circular for replication. |
|
|
Transformation Visual |
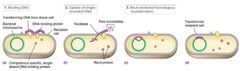
|
|
|
Def: Transduction |
Transfer of DNA from one cell to another by a bacteriophage. |
|
|
Two Modes of Transduction |
Generalized Transduction
&
Specialized Transduction |
|
|
Def: Generalized Transduction |
DNA from any portion of the host genome is packaged inside the virion.
Donor genes cannot replicate independently.will be lost without recombination. |
|
|
Def: Specialized Transduction |
DNA from a specific region of the host chromosome is integrated directly into the virus genome.
May be integrated during lysogeny, or homologous recombination may occur. |
|
|
Transduction |
Occurs in many Bacteria and at least one Archaea. Ex: multiple-antibiotic-resistance genes in Salmonella, Shiga-like toxins in Escherichia coli, virulence factors in Vibrio cholerae, photosynthetic genes in cyanobacteria. |
|
|
Generalized Transduction Pt. 1 |
Virtually any gene can be transferred to recipient (transductant). Sometimes host DNA accidentally packaged into phage, forming transducing particle that is defective and cannot lead to viral lytic infection. |
|
|
Generalized Transduction Pt. 2 |
Upon lysis, transducing particles and normal virions released. Recipients of transducing particles may recombine DNA. Low efficiency: one in 106–108 cells transduced. |
|
|
Lytic Cycle vs. Transduction |
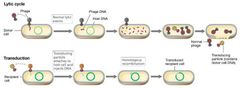
|
|
|
Lysogeny and Specialized Transduction Pt. 1 |
Extremely efficient transfer. Selective and transfers only small part of bacterial chromosome. Phage genome is integrated at specific site (e.g., Lambda in E. coli: next to galactose utilization genes). |
|
|
Lysogeny and Specialized Transduction Pt. 2 |
Viral replication is under control of bacterial host chromosome. Upon induction, viral DNA sometimes excises incorrectly and takes adjacent host genes along with it. Limit to amount of host DNA that can replace phage DNA, but helper phage can assist. |
|
|
Specialized Transduction Visual |
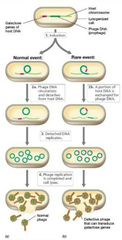
|
|
|
Phage Conversion Pt. 1 |
Alteration of the phenotype of a host cell by lysogenization. Prophage from normal, nondefective temperate infection becomes immune to further infection by same phage. Other phenotypic changes can also occur. |
|
|
Phage Conversion Pt. 2 |
Salmonella enterica serovar Anatum and bacteriophage ε15. Corynebacterium diphtheriae and bacteriophage β. Many natural lysogens found, suggesting this is an essential process for survival. |
|
|
Conjugation (Mating) Pt. 1 |
Horizontal gene transfer that requires cell-to-cell contact. Plasmid-encoded. Occurs between closely related or distantly related cells.
|
|
|
Conjugation (Mating) Pt. 2 |
Donor Cell: contains conjugative plasmid. Recipient Cell: does not contain plasmid. Other genetic elements (e.g., other plasmids or chromosome) may be mobilized (transferred during conjugation). |
|
|
F (“fertility”) Plasmid Pt. 1 |
~99 kbp circular DNA molecule. Contains genes that regulate DNA replication. Also contains transposable elements that allow the plasmid to integrate into the host chromosome. |
|
|
F (“fertility”) Plasmid Pt. 2 |
Contains tra genes that encode transfer functions (synthesis of sex pilus and type IV secretion system for DNA transfer). Pili allow specific pairing through receptor contact, pulling cells together, and DNA transferred through junction. |
|
|
F (“fertility”) Plasmid Visual |
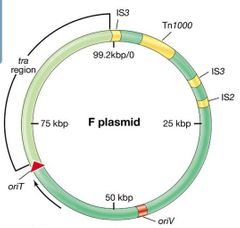
|
|
|
Mechanisms of DNA Transfer During Conjugation Pt. 1 |
DNA synthesized by rolling circle replication. Mechanism also used by some DNA viruses. Both donor and recipient have complete plasmids afterwards. |
|
|
Mechanisms of DNA Transfer During Conjugation Pt. 2 |
Transfer of plasmids is efficient and rapid; under favorable conditions nearly 100 percent recipients acquire plasmids. Transfer of F plasmid takes ~ five minutes. |
|
|
Formation of Hfr Strains and Chromosome Mobilization Pt. 1 |
F plasmid is an episome (can integrate into host chromosome). Cells possessing a nonintegrated F plasmid are called F+. |
|
|
Formation of Hfr Strains and Chromosome Mobilization Pt. 2 |
Cells possessing an integrated F plasmid are called Hfr (high frequency of recombination). High rates of genetic recombination between genes on the donor (Hfr) and recipient (F–) chromosomes. Integration provides mechanism for mobilizing a genome. |
|
|
Presence of the F Plasmid Results in Three Distinct Changes |
1) Ability to synthesize F pilus. 2) Mobilization of DNA for transfer to another cell. 3) Alteration of surface receptors so that cell can no longer be a conjugation recipient |
|
|
Integration of F Plasmid and Chromosome Mobilization Pt. 1 |
Insertion Sequences.
Homologous recombination results in F plasmid integration into chromosome.
After integration, tra functions normally and strain synthesizes pili. |
|
|
Insertion Sequences |
Present in both the F plasmid and E. coli chromosome providing regions of sequence homology. (IS; mobile genetic elements) |
|
|
Integration of F plasmid and chromosome mobilization Pt. 2 |
When recipient encountered, part of plasmid and chromosomal genes transferred.
Many Hfr strains possible. |
|
|
CRISPR |
CRISPR (Clustered Regulatory Interspaced Short Palindromic Repeats): A Prokaryotic “immune system” that evades viral destruction and maintains genome stability. |
|
|
CRISPR mechanism Pt. 1 |
Memory bank of incoming nucleic acid sequences for surveillance against foreign DNA.
Consists of segments of foreign DNA (spacers) that previously invaded cell alternating with identical repeated sequences. |
|
|
CRISPR mechanism Pt. 2 |
Key is transcription of long RNA molecule cleaved in the middle of repeated sequences by nuclease activity of CRISPR-associated (Cas) proteins.
Generates CRISPR RNAs (crRNAs) that base-pair with invading nucleic acids, resulting in destruction. |
|
|
Distribution of CRISPR |
Widely distributed (~90 percent of Archaea and 70 percent of Bacteria). Bacteriophages can overcome recognition by genome mutation. |

