![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
106 Cards in this Set
- Front
- Back
|
What is torsional stress in DNA? |
Coiling of closed circular DNA in 3 dimensions |
|
|
Linkng number of DNA? How is it calculated. |
Total count of times that two strands of circular DNA cross eachother(changed by supercoiling) Has + value for positive supercoils and - value for negative supercoils |
|
|
What is the function of supercoiling?
|
Some organisms (termophiles) use supercoiling to prevenmt DNA denaturation. DNA replication with negative supercoils as DNA is more relaxed |
|
|
What do "topoisomerase" enzymes do? How?
|
Increae or reduce the linking number. unwinds DNA.
|
|
|
Describe Type 1 Topoisomerase.
|
Change linking number by +/-1 by breaking one strand of DNA, passing other strand through the gap and sealing the gap. No ATP used.
|
|
|
Describe Type 2 Topoisomerase. Give an example.
|
Changes linking number by +/-2. Does so by breaking both strands of DNA and passing another part of dsHelix through the gap, sealing it afterwards. Ex: Gyrase
|
|
|
How is DNA organised in E.coli? What proteins maintain this organisation?
|
single circular DNA forms series of supercoiled loops radiating from central core protein. Much organised. Proteins: Gyrase(-2), Topoisomerase 1(+1) and HU proteins.
|
|
|
What is structure and function of HU proteins? numbers? |
Tetrameric structure that plays a role in DNA packaging just like histones in Eukaryotes. 60bp around one HU tetramer. ~13000 HU proteins HU - |
|
|
Types of Bacterial genomes? other than circular.
|
Some bacteria (agrobecterium tumefaciens, Streptomyces coelicolor) have LINEAR genomesSome bacteria have MULTIPARTILE genomes. (many circular molecules like plasmids etc.;)
|
|
|
what kind of genes plasmids usually carry?
|
usually plasmids don't have essential genes, they carry non-essential resistance, adaptation genes.
|
|
|
Variotion in prokaryotic genomes? How many BP per GENE?
|
in generral variation can be very bid depandant on species. Usuallu 1000bp ~ 1gene |
|
|
what is horizontal gene transfer?
|
Transfer of genes from organism to organism, not through generations!
|
|
|
Name and explain possible routes for horizontal gene transfer?. |
Prophages - viral DNA incorporated into bacterial genome , that can agg new genes(toxins) to bateria Genomic islands - parts of genome with horizontal transfer origins (acquired usually by transformation, transduction etc.) Transposable elements - transposons, jumping genes, can self excise and self insert into parts of genome. |
|
|
What does DNA pol III do?
|
its the main replicative enzyme that replicates DNA strands 5' to 3'
|
|
|
What does DNA pol I do?
|
Its replicative enzyme that removes primers from okazaki fragments by continuing DNA replication.
|
|
|
What does DNA ligase do?
|
Joins the strands of DNA during replication (on lagging strand.
|
|
|
What are Oazaki fragments?
|
Fragments of lagging strang that have RNA primers and some DNA on them. |
|
|
Define a REPLICON
|
Replicon - basic unit of replication. A sequence that has an origin of replication and MUST be replicated at least once per cell division
|
|
|
What is the link between REPLICATION and CELL DIVISION
|
DNA replication comits cell to undergo division Cell division cannot start unless replication has been completed |
|
|
Replicons in E.coli and origin of replication.
|
E.Coli has just one origin of replication (OriC), thus single replicon.
|
|
|
Direction or replication of DNA in E.coli.
|
DNA of E.Coli replicates in both direction of circle (bidirectional)
|
|
|
Describe structure of oriC, length of different parts. Can be drawn. |
minimum acting region is 245bp long. 14 GATC sequences 4-5 copies of 9bp right hand 2/3 of oriC 3 copies of AT-rich 13bp sequence left 1/3 part of oriC |
|
|
what is DNA melting
|
denaturation, unwinding, separation of strands
|
|
|
what bacterial toxins are of prophage origins?
|
Diphteria toxin, botulinum toxin, cholera toxin, shiga(food poisioning) toxin.
|
|
|
Describe in detail Initiation of replication in E.coli in prokaryotes. Can draw.
|
0) all 14 copies of A's in GATC must be methylated 1) DNA A binds to 9bp repeats = closed complex 2) 13bp regions MELT = open complex 3) then DNA B and C id loaded onto open complex 4) ATP is hydrolysed and DNA C released 5)DNA B is a helicase that unwinds strands bidirectionally. -single strand binding protein and Gyrase ait in this step. 6) primase synthesizes RNA primers on both strands an replication commences. |
|
|
What is he function of Single Strand Binding protein?
|
Holds DNA in single strands, prevents formation of basepairs. Cells immune system is also aains ssDNA, so SSB helps prevent immune response and allow succefull replication.
|
|
|
What type of molecule is DNA B
|
Helicase
|
|
|
What signals initiation of replication in E.coli?
|
Methylation of A in GATC sequences in oriC by Dam methylase.
|
|
|
What is Hemi-methylated DNA and how E.coli uses it during cell cycle
|
it is a half-methylated dna that is in the cell after replication. Hmi methylated DNA cannot replicate (ensures there is only 1 round of replication). this DNA is sequestered at a membrane site so cell can undergo cell division. (re methylation occurs in 1/3 of cell cycle)
|
|
|
Describe replication Termination event. Where does it happen? What is terminus region? What are traps and what they do? |
2 opposite replication forks fuse in the region opposite to oriC (Termination region). It has several traps that pause replication fork so it stands and waits for another. Termination only when anti-clockwise fork meets STALLED clockwise fork!
|
|
|
What is bacterial replication termination protein? How does it stoop replication fork?
|
Tus,binds to ter sites. It inhibits the helicase part of DNA pol 3
|
|
|
Which fork must be stalled for replication termination to occur? (Bacterial replication, E.Coli)
|
Clockwise
|
|
|
What is DNA recombination?
|
Breaking and joining DNA molecules in new fashion. produces mutations, rapid evolution, new gene combinations.
|
|
|
What are two types of recombination? Describe each
|
Homologous - sequence exchange between two similar(homologous) pieces of DNA like 2 homologous chromosomes. Non-Homologous - sequence exchange between non similar DNA molecules |
|
|
Types of NON-Homologous recombination?
|
Transposition (via Transposons) Site-specific recombination |
|
|
What is the significance of Homologous Recombination? |
Happens in all organisms and generates genetic diversity and DNA repair.
|
|
|
How long those homologous regions have to be for recombination to occur?
|
100-500bp
|
|
|
Describe the process(mechanism) of Homologous recombination?
|
1) Alignment - 2 Dna molecules become aligned 2) Cleavege - onestrand on each molecule gets cleaved at CHI site by RecBCD 3) Invasion 3' ends invade opposing molecule and form Holliday junction (RecA) 4) branch migration (holliday elongation) RuvAB 5) Isomerization - holliday strands cpontaneously cross and uncross 6) Resloutiuon -Holliday junction is cleaved by RuvC. Result depends on cleavege direction. |
|
|
Explain How RecBCD enzyme complex works?
|
RecBCD binds to DNA and starts unwinding and degrading both strands(23bp steps) it until encounters CHI site. When it reaches Chi its activity changes to 5' exonuclease, thus it leaves a 3' end sticking out(tail). RecA then binds to that tail stabilizing it and drivin invasion. |
|
|
What does RecA do?
|
it binds to 3' sticking out end after RecBCD and drives strand invasion |
|
|
What is Chi site on bacterial DNA?
|
It is the sequence at which RecBCD changes its activity to 5'to3' exonuclease. E.Coli has 1009 of them.
|
|
|
What are transposable genetic elements?
|
transposons, or jumping genes ar specific sequences that can transfer copies of themselves into other parts of the same or different molecule. They can also undergo inversion. |
|
|
What are Insertion sequences? how long?
|
Small (<2kb) long components of bacterial chromosomes. Only have transposition genes in them(don't carry phenotypical features). Have inverted repeats on ends and cause direct repeats at the site of insertion. |
|
|
Difference between Transposons and Insertion sequences?
|
Transposons usually encode a phenotypic characteristic, while Insertion sequences don't. IS are also shorter than transposons. |
|
|
Describe Tn3 Transposon. Length, What proteins does it encode, other specific sites. |
Tn3 is a 4957bp long transposon that encodes for 3 proteins: TnpA TnpR(transposition) and Bla B-lactamase (ampicillin resistance) |
|
|
Describe how Transposition in Tn3 occurs. (Replicative transposition)
|
The process is called replicative transposition. 1) a cointegrate with 2 transposon copies is created. (thus replicative). 2) molecule crosses over and recombines at 2 res sites. Leaves 2 molecules with transposon. |
|
|
What are Inversion sequences, and what function they do?
|
Inversion sequences are ones that cannot transpose, but can change their orientation within DNA molecule, co can control expression in some organisms.
|
|
|
What is hin region responsible for in Salmonella species?
|
It is the gene inversion of which controls 1 or phase 2 flagella expression.
|
|
|
Explain(Draw) regulationary circuit controlling Salmonella H1 and H2 flagella expression.
|
Most important part, its is controlled by HIN region that has a promoter to activate H2 and Rep - repressor.
|
|
|
What most phase-variable(invertable) are encoding for?
|
Surface proteins.
|
|
|
2 general classes of DNA damage in bacteria?
|
Single base changes - no effect on transcription/translation Structural distortions - impede transcription/translation |
|
|
5 types of DNA repair in prokaryotes. |
Direct repair - reversal or simple removal Mismatch repair - detection and repair of mismatched bases Excision repair - repair by excision of patch of DNA with replacement Tolerance systems - allow DNA replication through damaged regions Retrieval systems - using recombination to repair damaged regions |
|
|
Photolyase and direct repair.
|
Direct repair mechanism. 1)Photolyase binds to pirimidine dimers in the dark. 2)It has 2 chromophores that absorb light energy in the range of 300-600 nm which is used to split dimers apart. |
|
|
Mismatch repair in Prokaryotes
|
Ex: uracilDNA Glycosidase 1) UDG removes mismatched Uracil creating AP site 2) AP edonuclease then breaks phosphodiester backbone (closer to 5' site) 3) DNA pol 1 binds to the brek and synthesizes new patch of DNA which is sealed by DNA ligase |
|
|
How does mut system works?
|
Mut system (Proofeading) recognises mismatches ir insertion/deletions on hemi methylated DNA in prokaryotes. 1) MutS bind to mismatch. 2) MutL comes to stabilize it. 3) MutH is recruited to nearby methyl group and nicks UNMETHYLATED strand. 4) MutU heliceze unwinds DNA 5) DNA pol 1 and ligase do their job |
|
|
3 Types of excision repair in prokaryotes.
|
Very short patch: mismatches between basepairs Short patch: 20 nuleotides (99% of bulky lesions) Long patch: 1500-2000bp |
|
|
Explain bacterial excision repair mechanism.
|
1) enzyme uvrABC binds to damaged regions and cuts to the sides of them 2) UvrD helicase II separetes the strands 3) DNA pol 1 and DNA ligase repair the gap |
|
|
What are tolerance systems and how they work?
|
belong to Y-DNA pol family. Those systems use DNA polymerases that can bypass damaged DNA parts when synthesizing DNA. Ex: human Pol n can bypas UV caused pyrimidine dimers quite well |
|
|
What is special about Human DNA Pol n?
|
It can bypas UV induced pirimideine dimers, and usually inserts correct bases there. Significant role in Xeroderma Pigmentosum.
|
|
|
How do prokaryotes repair DNA using retrieval systems?
|
Retrieval systems repair DNA during replication. 1)The strand that has a mistake, gets a complimentary GAP.2) tha original(methylated) patch is inserted into that gap3) the new hole is resynthesized
* The damage is not repaired, but replication is done. "daughter strand gap repair" |
|
|
What is bacteria's SOS response an how is it activated? |
It is a mechanism that activates in case of severe DNA damage. allows error prone replication in expense of fidelity. RecA sences ssDNA(damage) and activates inhibiting editing by DNA pol 3 and cleaving LexA gene inhibitor. |
|
|
What is role of LexA in SOS response |
it is always active inhibitor of SOS response that binds to LexA box in promoter region inhibiting promoter |
|
|
Define stages if transcription |
Recognition - binding of RNA pol to promoter Initiation - creation of first ~9 nucleotide bonds and release of sigma factor Elongation - production of RNA 5' Termination - recognition of termination sequence and dissociation of enzymes |
|
|
Structure if bacterial RNA pol. roles |
(alpha)(beta)(beta') DNA binding, up element recognition (sigma)promoter recognition (omega) stabilization |
|
|
How does sigma factor influence RNA pol features. |
with this factor RNA pol is bad at binding to random DNA, but binds very well to PROMOTERS and melting DNA(1st closed then open complex) |
|
|
Describe the organisation of promoter sequence. |
2 Consensus sequences: -10 - TATAAT -35 - TTGACA |
|
|
What are UP and DOWN mutations (bacterial)? |
Mutations occurring in consensus sequences making it more similar to consensus(UP) or less similar(DOWN). can make promoter stronger or weaker (alter binding affinity) |
|
|
-35 and -10 DOWN mutations |
-35 DOWN mutations prolong the CLOSED complex formation(recognition) -10 DOWN mutations prolong OPEN complex formation (melting) |
|
|
What is an up element? How is it recognised? |
UP element further increases promoter strength, is ~20bp upstream of promoter. Consists of rich AT region Is recognised by (alpha) subunit |
|
|
What determines promoter strength? |
-10 sequence -35 sequence UP element |
|
|
What is the role of (sigma) factor? |
Recognises and binds to the promoter region, initiating formation of first ~9 phosphodiester bonds. It later dissociates making DNA pol more friendly to non-promoting regions and allowing elongation. There can be different (sigma) factors recognising different promoters. |
|
|
Describe replication initiation? |
RNA pol sigma subunit creates high affinity for promoter. RNA pol binds to promoter and closed complex is formed. DNA melts (open complex) and first 9~ nucleotides are bound. then sigma dissociates and Elongation starts |
|
|
Define CONSTITUTIVE GENE EXPRESSION
|
genes that are always on, constantly transcribed.
|
|
|
Define an OPERON
|
Clusters or genes that are controlled by single promoter(as if they were a single gene), thus transcribed together.
|
|
|
What is polycistronic mRNA.
|
mRNA that produces multiple proteins(linked by their function).
|
|
|
Repressors and activators. Inducers and co-repressors. whats the difference |
repressors and activators - are proteins that bind to DNA and either activare or repress transcription Inducers and Co-repressors - act on proteins not genes |
|
|
Define REGULON
|
Many operons that are controlled by single regulatory protein
|
|
|
what is GLOBAL REGULATION SYSTEM(bacterial transcription)
|
A major transcription regulation system that has a very wide control range usually activated by single environmental factor (e. g. glucose depletion)
|
|
|
What is Diauxic growth?
|
Bacterial growth using 2 different carbon sources.
|
|
|
Describe structure of lac operon |
(lacI)(CAP site)(promoter)(operator)(lacZ)(lacY)(lacA) lacI - inhibitor that is always expressed CAP site - cAMP activator protein Promoter(lacP) - where RNA pol binds Operator(lacO) - Where lac repressor binds lacZ - encodes beta-galactosidase lacY - encodes beta-galactoside permease lacA - encodes beta-gaactoside transacetylase(detoxifying role) |
|
|
What carbon sources induce/repress lac operon
|
induction by lactose reression by glucose |
|
|
Describe precise lac operon induction. |
1) repressor inactivation by lactose -low levels of beta-galactosidase are present in cell(leaky expression) -if lactose is present it becomes allolactose -allolactose binds to LacI causing allosterical shape change that causes dissociation of repressor from operator 2) CAP activation(glucose deficit) -normally glucose blocks adenylate cyclase activity -when glucose is absent, cAMP is produced, that binds to CAP and activates it -CAP binds to CAP site and recruits RNA pol |
|
|
Why is CAP protein needed at all? why not just inhibitor?
|
This is because RNA pol has low affinity to lac operon promoter as its quite different from consensus sequence. Thus CAP needed to recruit it.
|
|
|
CAP and RNA pol interaction
|
7 AA beta-turn of CAP interacts with alpha-subunit of RNA pol
|
|
|
How was lac operon regulation discovered? |
by Monod and Jacob. Using merodiploid(partially diploid) E.coli. With 2 lac operons on different DNA molecules and mutating them to see if they are complementary. Found cis- and trans acting- elements |
|
|
Explain gene complementation using example of 2lac operons in merodiploid cell
|
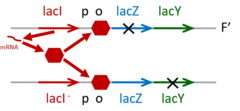
|
|
|
Characterize lac mutations. lacZ- Haploid phenotype lacY- Dominant/recessive lacA- cis/trans acting LacI- LacOc lacP- lacIs lacI-d |
kevin purdy L3. check answers on moodle. |
|
|
Define cloning and cloning vector? |
Cloning vector - a plasmid or a piece of DNA inserted into virus that cam be transferred to living cells ad then replicate in them Cloning - a successful transfer of desired DNA fragment to bacteria that will replicate and pass to next generations, giving them phenotypic traits. |
|
|
Describe DNA cloning into bacteria process? |
Firstly take a plasmid and using restriction enzumes insert desired DNA into it. Transfect cloning vector into bacteria (E.coli) by bacterial transformation(heat shock, electrick chock) or viral pransduction (lambda phage, P2 phage). Culture bacteria. Select Vector bearing bacteria by using selectable marker (antibiotic resistance, metabolic phenotype).
Lysing cells to release cloning vector can be done. |
|
|
Cloning RNA. Where is it useful? |
RNA cannot be cloned directly. Take mRNA, use RT to make DNA out of it. Then clone DNA.
Useful in analysing eukaryotic genes with introns as mRNA deletes introns. Can see which parts of genes are introns, exons. |
|
|
2 Types of Lambda vectors Insertion vectors Replacement vectors Lambda vector capacity? |
Insertion vectors - bit of phage non-essential DNA has been removed to make restriction site. New DNA can be inserted into that restriction site Replacement vector - a bit of non-essential phage DNA is replaced with non coding "stuffer" that then can be replaced with desired DNA Capacity: 20kbp |
|
|
Fosmids and cosmids capacity |
Fosmids are large plasmids based on bacterial F' plasmid. Cosmids are plasmids that contain cos sequence that allows them to be packed into lambda phage Capacity 40kbp After transfection are maintained in cell as plasmids. |
|
|
Ti plasmids. |
Ti plasmids are found in plant bacteria Agrobacterium tumafeciens (pathogen). A cmall portion of Ti plasmid is transferred into the plant cell and is then integrated into plant genome. Can be used for GMO plants. |
|
|
Artificial chromosomes |
Bacterial artificial chromosome(BAC) is based on F' plasmid and can carry up to 300kbp. Used to investigate genome regions outside their hosts. Yeast artificial chromosomes(YAC) - based on real yeast chromosomes. Must have ori's, Centromere and telomeres(usually are circularised). Usually have selectable and screenable markers on both arms, so only cells having full chromosome can grow. Can carry 600kbb. Mega-YAC's can carry 1.5Mbp |
|
|
Selectable marker vs screenable marker |
Selectable marker - gene that gives trait suiteble for artificial selection (antibiotic resistance, auxotrtophic) Screenable marker(reporter gene) - genes that make transfected cells look differently, usually cause colour change that allow to visibly see which cells have been successfully transfected. (GFP) |
|
|
Desciribe addiction cassette RNA regulation. Death protein. |
RNA regulation is regulation via small non-coding RNA molecules. Ex: 1)Cassette produces stable Lethal protein mRNA and unstable anti-sense RNA. 2) ncRNA binds and prevents death protein production. 3) If cell without plasmid is produced. then ncRNA cannot be made, so it is degraded and lethal protein is made. |
|
|
Describe iron use regulation in E.coli by RNA |
If iron is available cell produces iron requiring proteins. FUR(Fe) represses ryhB. If iron becomes limited, Fe dissociates from FUR and fur stops ryhB repression. ryhB ncRNA is made that goes and degrades mRNA's of iron requiring proteins. = cell requires less iron. Regulated by iron concentration. |
|
|
Examples of positive and negative interactions in microbial communities. |
Positive - toxic metabolite of one product may be substrate for another. Cyanobacteria live longer in presence of heterotrophs. Negative - competition, antibiotic production. |
|
|
What is quorum sensing? |
Ability of bacteria to sense their density and abundance. Bacteria secrete autoinducers and have receptors for them. Each cell produces too little autoinducers to activate receptors, but when many cells are present autoinducer concentration rises, reaches treshold and results in positive feedback loop. This then changes gene expression. |
|
|
Quorum sensing and Staphylococcus aureus. |
When there are little cells - produce biofilm (AGR regulatory system) AgrD - Autoinducing peptide AgrB - Transmembrane protein secretes AIP AgrC - Recetor that senses AIP and phosphorylates AgrA AgrA - Response proteins P3 producing RNAIII
As density of cells rise, concentration of AIP rises. AgrA gets phisphorylated and RNAIII made
RNAIII acts as mRNA for hemolysinDelta and Regulates many genes - represses adhesins, induces proteases, capsule toxins, AlphaHemolysin(RNA binds to shine delgano region of mRNA hlaA activates translation).
DRIVES INVASION |
|
|
What are biofilms? Why do bacteria need it? |
Structured clusters of cells coated in polymer matrix and attached to surface (cells, ticcues solid surfaces like implants) Allows pathogen survival and proliferation in environment and hosts. Protects against immune response, dessication.
|
|
|
Common characteristics of biofilms? |
Polymer matrix coating cells(exopolysacharides, proteins, nucleic acids) Formation initiated by extracellular signals |
|
|
5 stages of biofilm development? |
1) initial attachment (with flagella, type 1 pili) 2) Irreversible attachment (with LPS, type 4 pili) 3) formation of microcolonies (repress flagella, produce alginate) 4) maturation 2 - Quorum sensing 5) Dispersion - burst and release of planktonic cells |
|
|
Multispecies biofilms - why are they beneficial? |
Set of positive interactions allows biofilm to function like small ecosystem, promoting bacterial survival |
|
|
Genes in biofilm maturation? |
switch gene expression and protein production for biofilm maturation. -Environmental signal for flagella switch in Vibrio parahaemolyticus -sigma22 factor that changes protein expression in Pseudomonas aeroginosa to maturate into microcolonies. |
|
|
What regulates biofilm formation? |
Quorum sensing in staphylococcus aureus Surface sensing by Vibrio parahaemolyticus Changing gene expression using sigma factor. |

