![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
27 Cards in this Set
- Front
- Back
|
At which 6 steps can eukaryotic gene expression be controlled?
|
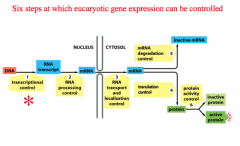
1. Transcriptional control
2. RNA processing control 3. RNA transport and localization control 4. Translation control 5. mRNA degradation control 6. Protein activity control |
|
|
DNA-Binding Protein Motids
|
DNA-binding proteins often contain motifs or "Super-Secondary Structures" which are compact 3-D collections of several secondary structures (alpha helices, beta sheets).
These super-secondary structures serve an identifying purpose as they can be used to identify or predict the function of a protein. For example, we can predict that a protein will bind DNA if it contains a helix-turn-helix motif. |
|
|
Helix-turn-helix
|
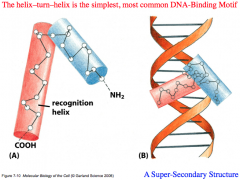
The simplest, most common DNA-binding motif.
Super-secondary structure comprised of 2 alpha helices joined by a short sequence of amino acids. The alpha helices contain R groups which project outward and are able to interact with DNA. One helix functions as a "Recognition Helix" because it interacts in a sequence-specific way with the major groove of DNA. The second alpha helix is a "Structural Component" that helps to position the recognition helix. Usually function as dimers, and are capable of binding palindromes equally on both sides of DNA |
|
|
Zinc Finger
|
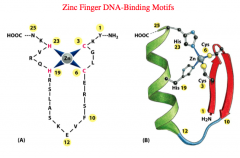
In this DNA-Binding Motif, cystidine or histidine residues chelate Zinc residues and form a finger of amino acids. Zinc stabilizes the finger-like projection of amino acids and enables an alpha helix in the projection to interact in the major groove of DNA.
Zinc fingers tend to be dimers or large proteins with two different binding domains. The glucocorticoid receptor has two subunits and uses a zinc finger to bind DNA effectively. |
|
|
Leucine Zipper
|
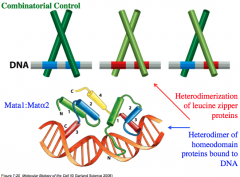
DNA Binding Motif
Leucine zippers form in DNA-binding proteins where there is a leucine residue present every 7th amino acid. This results in leucine residues all along the same side of the alpha helix since every 7th amino acid places the R group of Leucine on the same side of the alpha helix. These leucine-rich alpha helices can assume a hydrophobic domain, which excludes water and encourages the two alpha helices to associate tightly, forming effective DNA-binding domains. Many dimers assume homodimer or heterodimer leucine zippers that enable them to interact with the major groove of DNA. |
|
|
DNA Binding Proteins and Regulation of Gene Expression
|
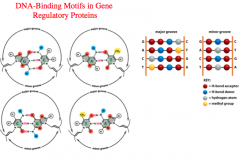
DNA Binding Proteins need access to the primary sequence of DNA that contains the information in order to decide which genes need to be transcribed.
But, it would be pretty inefficient to unwind the DNA all along the genome to find a sequence, so instead they have a way of being able to read the DNA sequence's information on the outside while it is in the helical formation In the Major Groove, we can distinguish base pairs from each other and what the sequence is on each strand. But, in the Minor Groove, we can only distinguish sets of base pairs, but not what the overall sequence is. *Because the major groove is more open, DNA binding proteins get a better look at the DNA sequence. Therefore, it is common for DNA binding proteins to interact with the major groove of DNA.* -These proteins can recognize consensus sequences for restriction sites (sites to cut), promoter elements, or enhancer or repressor sequences in the DNA. |
|
|
What facilitates the interaction of a DNA binding protein and the major groove of DNA?
|
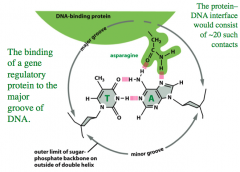
Facilitated by the R groups of the primary sequence of amino acids on the protein.
These contact the DNA and form any number of weak molecular forces. For example, Asn forms H bonds with DNA. These are linear and sufficiently close and tight fitting bonds to exclude water. The sum of all these contacts makes a very tight interaction. The protein-DNA interface would consist of ~ 20 such contacts. |
|
|
Genetic Switches: The trp Operon (overview)
|
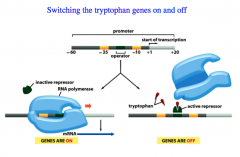
The switch turns on and off as a function of nutrient availability.
The operon contains the enzymes necessary for tryptophan (trp) biosynthesis. The bacterium will turn this ON when Trp is ABSENT from the environment, and OFF when Trp is PRESENT. Clever way of conserving resources when it isn't necessary to synthesize tryptophan. |
|
|
How does the trp Operon work?
|
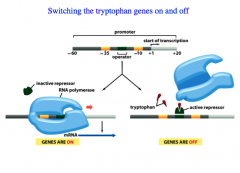
RNA Polymerase binds to a regulatory sequence.
Inside the promoter region, there is an Operator Sequence, and the Repressor recognizes this sequence. Trp is an allosteric modifier of the repressor. -When Trp is present, it binds to the repressor and enables the repressor to bind to the operator sequence, preventing RNA polymerase from binding. (This prevents the genes that encode for Trp biosynthesis from being transcribed) -When Trp is absent, the repressor is NOT activated and thus does not bind to the operator sequence, and allows RNA Pol to bind to the promotor region. (The genes that encode for the biosynthetic pathway ARE transcribed). *Trp present: repressor ACTIVE, genes NOT transcribed *Trp absent: repressor INACTIVE, genes ARE transcribed |
|
|
Negative Regulation
|
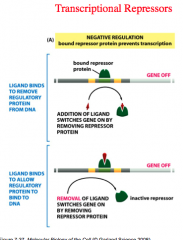
negative regulation means that there is a bound repressor protein that prevents transcription
|
|
|
Positive Regulation
|
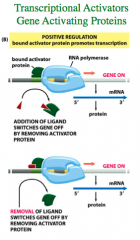
Positive regulation means that a bound activator protein promotes transcription.
|
|
|
Position Effects of Regulator
|
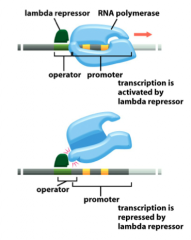
Some bacterial gene regulatory proteins can act as either a transcriptional activator or a repressor, depending on the precise placement of their DNA-binding sites.
Position of the regulator can be affected simply by the relationship between the DNA binding site for the protein and the RNA polymerase binding site In the bottom image, we see that by moving the repressor over by just one base pair, it now interferes with RNA polymerase binding and will block the binding site |
|
|
Lac Operon (overview)
|
Regulates the genes that are necessary for lactose catabolism. Again, it wouldn't make sense to expend the energy to synthesize these enzymes when lactose is not present. Moreover, the bacteria would prefer to use glucose as an energy source, and thus this system should be repressed when glucose is available
|
|
|
How does the Lac Operon work?
|
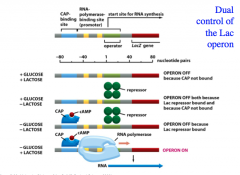
There is a dual control of the Lac operon because glucose and lactose levels control the initiation of transcription of the Lac operon via CAP (an activator protein) and the Lac repressor protein.
Both the presence of CAP and the absence of Lac repressor are required for the Lac operon to function. When lactose is present, it will bind to the repressor protein and inactivate it so that the repressor cannot bind to the operator. When glucose is absent, this leads to an increase in cAMP levels. The activator protein CAP can bind cAMP and activated CAP-cAMP can bind to the CAP-binding site on DNA. THUS, only when there is LACTOSE PRESENT and GLUCOSE ABSENT will the lac operon be activated. -this is because the repressor will be inactivate (due to the presence of lactose) and CAP will be bound (due to the absence of glucose) There are four possible situations (see image) |
|
|
Functional Domains on Transcription Factors
|
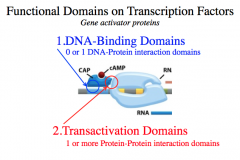
1. DNA-Binding Domains (0 or 1)
-for DNA-Protein interaction domains 2. Transactivation Domains (1 or more) - Protein-Protein interaction domains *Functionally independent of each other!* The major function of transcription factors is to help regulate transcription through multiple protein-protein interactions (via transactivation domains), which creates the transcription initiation complex. |
|
|
the modular strutter of a gene activator protein
|
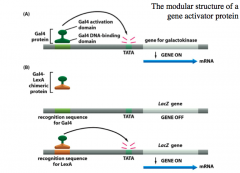
The DNA-binding domain and Transactivation domain(s) of transcription factors tend to be compartmentalized and functionally independent.
-We illustrated this with experiments wight he Gal4 transcription factor. The Gal4 transcription factor has a single DNA-binding domain and a single transactivation domain, and drives the expression of the galactokinase gene. If we replace the Gal4 DNA-binding domain with the DNA-binding domain for LexA, the Gal4-LexA chimeric protein cannot bind to the Gal4 DNA recognition sequence and the galactokinase gene will be turned off. However, if we replace the DNA recognition sequence with the LexA sequence, there is normal galactokinase expression since the chimeric Gal4-LexA protein can bind to the LexA DNA sequence (vie LexA DNA-binding domain) and the GAL4 transactivation domain can drive galactokinase gene expression. *you must have the appropriate DNA binding sequence for a DNA-binding domain to function* |
|
|
Promotors
|
DNA Elements that Regulate Gene Expression
Promotors establish *minimum conditions* for baseline transcriptional activity -cant get proper promoter function without enhancer -are position and orientation dependent because of their need to convey direction, therefore NOT palindromes! |
|
|
Enhancers
|
DNA Elements that Regulate Gene Expression
Enhancers *regulate* transcriptional activity. -Sequences of DNA that convey either positive or negative control over transcription. Features: -Do not contain promotor function -May exist outside of promotor region (Position Independent)* -Orientation independent (almost always exist as Palindromes)* Do NOT convey direction!! |
|
|
Two kinds of Transcription Factors that bind to enhancers
|
1. Constitutive Transcription Factors
-not regulated 2. Inducible Transcription Factors - can be turned on and off - Three sub-types: Second Messenger-Dependent, Hormone Receptors, and Tissue-Specific |
|
|
Genetic Control Regions
|
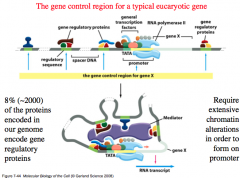
Genetic control regions get pretty big because multiple activators and repressors can combine with each other and bind to form a large complex.
The goal is to create an inviting environment for RNA Polymerase* -Or uninviting if you want to inhibit gene transcription. But this complex will not fit under normal conditions and thus this complex requires extensive chromatin alterations in order to form on the promoter. |
|
|
Four mechanisms that eukaryotic activator proteins use to direct local alterations in chromatin structure to open it up for transcription initiation
|
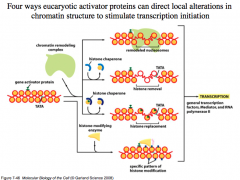
1. General activator proteins can recruit a chromatin remodeling complex to open up the DNA.
2. They can recruit histone chaperones that remove histones. 3. They can recruit histone chaperones that replace histones (these may not coil as much, or not associate with each other as well). 4. Modify the histones (via acetylation, methylation, etc.) to alter their properties. |
|
|
The Human Interferon Gene: The Histone Code and Transcription Initiation
|
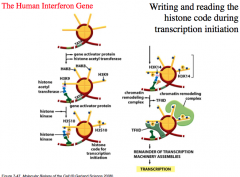
Illustrates how activator proteins alter chromatin structure.
1. Gene activator protein binds to DNA packaged into chromatin and recruits a histone acetyl transferase to acetylate lysine 9 of histone H3 and lysine 8 of histone H4. 2. Only after lysine 9 has been acetylated, a histone kinase is attracted by the gene activator protein and phosphorylates serine 10 of histone H3. 3. Serine modification signals the histone acetyl transferase to acetylate position K14 of histone H3. 4. This complex can then recruit chromatin remodeling complex and TFIID, which can bind and promote transcription initiation. Each step sequentially recruits modifying proteins and leads to a new interface for the recruitment of another protein and this forms a binding site for a larger complex (chromatin remodeling protein) to ultimately create an inviting initiation site and promote transcription initiation. |
|
|
Order of events leading to transcription initiation of a specific gene
|

order can vary between genes but you should know the events that occur
|
|
|
gene activator proteins work _______
|

synergistically!
there are gradients of expression depending on combinations that make up the complex. |
|
|
6 ways in which eukaryotic gene repressor proteins can operate
|
1. Competitive DNA Binding
-Gene activator and repressor proteins compete for binding the same regulatory DNA sequence (overlapping binding sites) 2. Masking the activation surface -repressor protein binds to and blocks the transactivation domain of a gene activating protein. 3. Direct interaction with the general transcription factors -repressor prevents the assembly of general transcription factors, which cannot interact with the initiation complex and RNA Pol. 4. Recruitment of chromatin remodeling complexes -repressor recruits a chromatin remodeling complex, which returns the nucleosomal state of promoter to pre-transcriptional form 5. Recruitment of histone deacetylases -repressor attracts a histone deacetylase to remove acetyl groups (which normally promote transcription initiation). 6. Recruitment of histone methyl transferase -repressor attracts a histone methyl transferase to methylate histones, which maintains chromatin in a transcriptionally silent form. |
|
|
Architectural Proteins
|
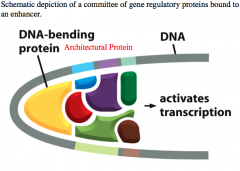
binding proteins which bend the DNA.
This allows other proteins bound to enhancers far from the promoter to interact with the transcriptional machinery at the promoter region. Some DNA sequences lend themselves to being hinges/turns in the DNA to enable this type of bending. |
|
|
Insulators and Barrier Sequences
|
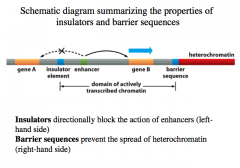
In highly regulated DNA, we do not want an enhancer sequence for one gene impacting a nearby genes.
So we have Insulators, which are DNA sequences that directionally bloc the action of enhancers. Insulators function by binding proteins and preventing enhancer sequences from acting beyond the gene of interest. Barrier Sequences function as fences to prevent spread of heterochromatin, heavily packaged and silent DNA, which could turn off a gene if it moved into an active gene. Insulator= Upstream Barrier= Downstream |

