![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
49 Cards in this Set
- Front
- Back
|
What is DNA metabolism? |
Consist of tightly regulated processes that achieve: A new copy of dna to fidelity before each cell division Repairs made to errors in dna synthesis Novel dna created through Recombination (segments of 2 dna molecules rearranged) or rearrangement within chromosome |
|
|
3 rules of dna replication |
1. Replication is semiconservative 2. Begins at an origin and proceeds bidirectionally (usually) 3. Synthesis of new dna occurs in 5-3’ direction and is semi-discontinuous |
|
|
Semi-conservative replication |
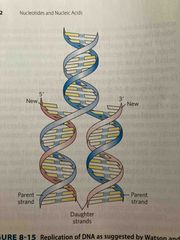
Meselson-stahl experiment: N is divided equally between 2 daughter genomes So each new strand has one parent and one new daughter strand |
|
|
Meselson-stohl experiment |
Used Heavy N (15) for one strand and light N (14) for another and allowed cells to divide Found one heavy and one light (parent and daughter) strand Allowed cells to divide again and had 2 of the hybrid plus 2 that were all light |
|
|
DNA is bidirectional (cairns experiment) |
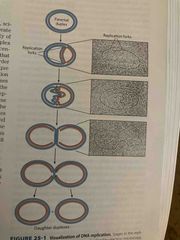
Showed in bacterial dna that both strands replicated simultaneously 2 replication forks suggested bidirectional replication |
|
|
Can replication begin anywhere? (Inmans experiment) |
Found dna always started at A-T rich regions forming bubbles. These bubbles were mapped (denaturation mapping) and found loops initiated at unique points and called them origins |
|
|
Leading vs lagging strand |

Synthesize dna in 5->3’ direction Because one parent strand is in 3->5’ direction, it is easier to replicate and is called the leading strand (does it faster so leads) and is continuous The other parent strand is in 5->3’ so the daughter strand is made in small segments to overcome which takes time (lagging “behind” strand) and is discontinuous due to Okazaki segments
|
|
|
Enzymes that degrade dna segments |
Nucleases degrade nucleic acids (DNases degrade dna and RNases degrade rna) Exonucleases cleave bonds that remove nucleotides from ends of dna Endonucleases cleave bonds within DNA sequence |
|
|
Enzymes that create dna segments |
Polymerases DNA polymerase I was discovered by kornberg |
|
|
Chemistry of dna elongation |
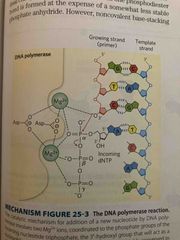
Nucleoside triphosphates serve as substrates in strand OH acts as a nuclephile It is activated via Mg by deprotonating the OH group making it a better nucelophile The OH attacks the alpha phosphate at the 3’ end of the growing chain Pyrophosphate is a good leaving group |
|
|
DNA polymerase requires a primer |
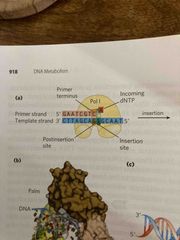
A short strand complementary to the template Contains a 3’ OH to begin the first DNA polymerase-catalyze rxn Can be made of dna or rna (more common) |
|
|
DNA polymerase |
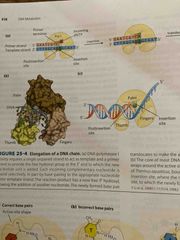
Enzyme that has a pocket with 2 regions: Insertion site- where incoming nucleotide binds Postinsertion site- where the newly made base pair resides when the polymerase moves forward |
|
|
Processivity |
The number of nucleotides added by DNA polymerase before it dissociates Processivity varies among DNA polymerases (each has its own and each also has its own polymerization rate) Can vary from a few to thousands |
|
|
Geometry of base pairing accounts for high fidelity |
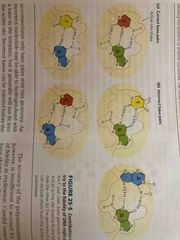
DNA polymerase active site excludes pairs with incorrect geometry Sometime will insert wrong one but has ways to fix these errors |
|
|
Errors during synthesis are corrected by 3->5’ exonuclease activity |
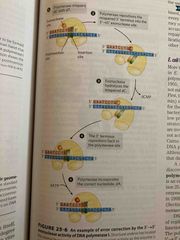
All DNA polymerase have an additional activity Exonuclease “proofreads” synthesis for mismatch base pair Translocation of enzyme to next position is inhibited until enzyme removes incorrect nucleotide |
|
|
In e. Coli there are at least 5 dna polymerases |
DNA polymerase I is abundant but not ideal for replication bc it has low processivity It’s primary function is clean up DNA polymerase III is the replicator II, IV, and V are involved in dna repair |
|
|
Klenow fragment |
A distinct domain that can be separated by protease tx DNA polymerase I has additional 5->3’ exonuclease activity that moves ahead of the enzyme and hydrolyze a things in its path Nick translation-a strand break moves along with enzyme |
|
|
Nick translation |
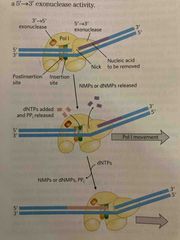
A break in strand Replace a segment |
|
|
DNA polymerase III complex |
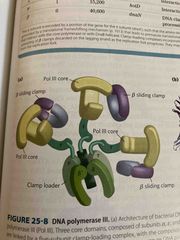
Has: 3 cores with beta subunits-beta subunits increase processivity of the complex Clamp loader- clamps on dna and then feeds through cores |
|
|
Replisome |
A set of 20 enzymes and proteins that are needed for dna replication in e.coli Includes: Helicases-use atp to unwind dna strands Topoisomerases- relieve stress caused by unwinding DNA-binding proteins- stabilize strands Primase- make rna primers Dna ligases- seal nicks btw Okazaki fragments |
|
|
Initiation of replication in e.coli |
Begins at oriC site A-T rich region |
|
|
Proteins and steps in initiation of replication in e. Coli |
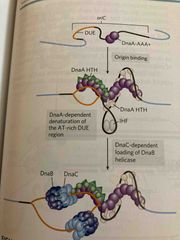
DNA Wraps around dnaA protein complex forming a supercool which leads to denaturation of DUE (dna unwinding element) sites DnaB migrates along 5-3’ and unwinds helix (helicase) Other proteins link to dnaB Single-stranded dna binding protein (SSB) stabilizes strands DNA gyrase relieves tension ahead of forks |
|
|
Regulation of initiation via methylation |
Dam (dna adenine methylase) methylase hemimethylates oriC so it can interact with plasma membrane Allows new dna to bind |
|
|
Elongation of leading strand |
Straightforward approach Primase (dnaG) makes rna primer (interacts with helicase but moves in opposite direction) DNA polymerase III adds nucleotides to the 3’ end of the strand |
|
|
Elongation of the lagging strand |
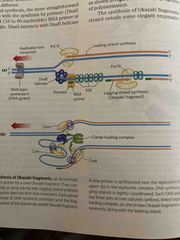
RNA primer and DNA polymerase III are the same Synthesis proceeds with nucleotides adding 3’ end Elongated away from fork One DNA polymerase III dimer complex synthesizes both strands |
|
|
Leading and lagging synthesis |
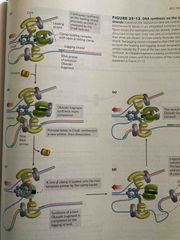
Core subunits of DNA polymerase III dissociate from on beta clamp and bind a new one RNA primer is removed by DNA polymerase I or rnase H1 DNA polymerase I fills the gap DNA ligand seals backbone |
|
|
3 rd stage of replication is termination |
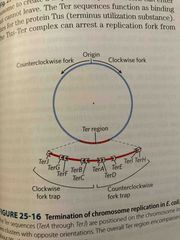
Replication forks meet at Ter sites found near each other in opposite directions Ter is binding site for Tus (terminus utilization sequence) that cause a replication fork to stop |
|
|
Replication in eukaryotes |
More complex than bacteria Yeast have 400 origins so called autonomously replicating sequences (ARS) Entire genome replicated 1x/cell cycle Regulation due to cyclin dependent kinases (CDK) which are ubiquinated for destruction at end of mitosis phase
|
|
|
Initiation of replication in eukaryotes |
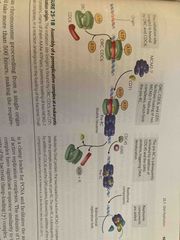
Require a pre-replication complex (pre RCs) Origin recognition complex (ORC) loads a helicase onto DNA (like bacterial dnaA) Helicase is a hexamer of mini chromosome maintenance protein (like dnaB) Occurs more slowly than bacteria (50 nucleotides/sec) |
|
|
Multiple DNA polymerases in eukaryotic replication (3 primary mulitsubunit) |
1. DNA polymerase alpha: has primase activity in one subunit and polymerization in another Does not have 3-5 proofreading 2. DNA polymerase E: synthesizes leading strand Highly processive and has proofreading 3. DNA polymerase &: Synthesis for lagging strand and has proofreading
|
|
|
Termination of replication in eukaryotes |
Synthesis of telomeres (found at ends of chromosomes) Enzyme telomerase uses rna |
|
|
DNA repair and mutations |
Damage to dna genome -majority are fixed but some escape repair changing daughter dna Accumulations of mutations is correlated to cancer There are 1,000 of lesions daily (Unrepaired dna) but only 1/1000 lead to mutations Human genome contains genes for over 130 repair proteins |
|
|
DNA lesions and mutations |
A lesion is dna damage If unrepaired, leads to mutation Mutations can be substitutions (point mutations), deletions, additions, and silent mutations (have no effect on gene functioning) |
|
|
Type is dna damage |
Mismatch (from incorrect nucleotides) Abnormal bases from deamination, alkylation, free radicals Formation of pyrimidines due to UV light exposure Backbone lesions from radiation and free radicals |
|
|
Mismatch repair in e. Coli |

One parent strand is methylated ( DAMmethylase) insert CH3 The new daughter strand is unmethylated for a short period of time after synthesis (any errors will reside here) The methyl -directed mismatch repair system will cleave the unmethylated strand in the initial part of the repair process |
|
|
Methylation-directed repair in e.coli |
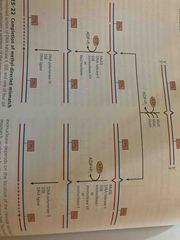
See pic |
|
|
Base excision repair |
Uses dna glycosylases (recognize specific lesions and cleave between sugar and base) Uracil glycosylase removes uracil (C deaminates to U and U is in RNA) |
|
|
Base excision repair in depth |

Nucleotide or region is removed by AP endonucleases Then DNA polymerase I adds new dna DNA log see seals Nick |
|
|
Nucleotide excision repair |

Lesions include: pyrimidine diners, photoproducts (UV light damage for both of those), benzopyrenguanine (from cigarette smoke) Removal of dna by excinucleases |
|
|
Direct repair includes: |
Photolyases (but don’t have this) O6-methylgaunine-dna methyltransferase (repairs methylated gaunine) AlkB demethylates 1-methylase nine and 3- methylcytosine |
|
|
If there is no undamaged dna to use as a template, we use |
Another chromosome (recombination) Or Error-prone translesion synthesis (TLS) |
|
|
Error prone tranlesion |
In bacteria: sos response when dna damage is extensive (involved DNA polymerase V) In mammals: TLS polymerases that recignize specific types of damage and respond (dna n when t-t dimer halts fork) Unmutable |
|
|
DNA recombination |
Segments of dna can rearrange within chromosomes from one to the other Involved in repair of dna, segregation of chromosomes during meiosis, and increase in genetic diversity Driving force of evolution along with mutations Helps virus and bacteria develop resistance |
|
|
3 classes of recombination |
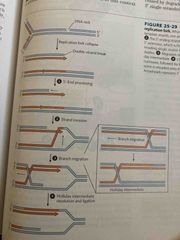
Homologous: exchange btw dna that share a region of similar sequence Site-specific: exchange only at a particular sequence DNA transposition: jumping genes |
|
|
Homologous replication in eukaryotes |
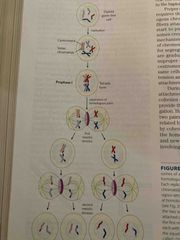
Occurs frequently during meiosis Crossing over in first meiotic division at chiasmatas where dna breaks and rejoins (called hot spots also) At the end of meiosis II: sister chromatids are separated with different genetic material inceasing genetic diversity |
|
|
Crossing over/ recombination |
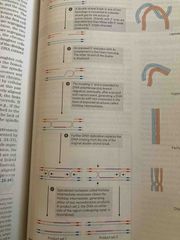
See pic |
|
|
Nonhomologous end joining |
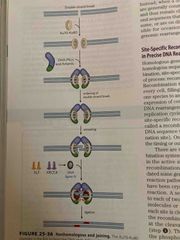
Another way to repair Not ideal Broken chromosomes ends are lighted back together No conservation of DNA sequence |
|
|
Site-specific recombination |

Limited to specific sequences with either Ter or Ser Recombinase cleaves strand and joins with new partners |
|
|
Transposable genetic elements (transposons) |
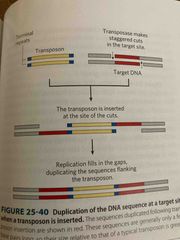
DNA that carries genes for transposases can move to a nee region or chromosome Cut themselves out and insert elsewhere and then these sites can be duplicated Ie: develop antibiotic resistance |

