![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
23 Cards in this Set
- Front
- Back
- 3rd side (hint)
|
What is recombinant DNA technology and why is it done? |
Involves transferring a fragment of DNA from one organism to another. The transferred DNA can be used to produce a protein in the cells of the recipient organism. |
|
|
|
Why does Recombinant DNA technology work even when the recipient and donor organisms aren’t from the same species? |
• the genetic code is universal (same codons code for same amino acids in all living things) • transcription and translation mechanisms are fairly similar in all living things |
|
|
|
In order to transfer a gene from one organism into another, you first need to get a DNA fragment that contains the target gene. Name the 3 ways that DNA fragments can be produced? |
1. using reverse transcriptase 2. using restriction endonuclease enzymes 3. using a gene machine |
|
|
|
Describe how DNA fragments are produced using reverse transcriptase? |
1. mRNA (complementary to the target gene) is isolated from cells 2. Mixed with free DNA nucleotides + reverse transcriptase 3. Reverse transcriptase uses the mRNA as a template to synthesis new strands of cDNA (complementary DNA) (The cDNA is a complementary copy of the mRNA due to specific base pairing) |
|
|
|
Describe how DNA fragments are produced using reverse transcriptase? |
1. mRNA (complementary to the target gene) is isolated from cells 2. Mixed with free DNA nucleotides + reverse transcriptase 3. Reverse transcriptase uses the mRNA as a template to synthesis new strands of cDNA (complementary DNA) (The cDNA is a complementary copy of the mRNA due to specific base pairing) |
|
|
|
What is the principle behind producing DNA fragments using reverse transcriptase? |
• most cells only contain 2 copies of each gene - difficult to obtain a DNA fragment containing the target gene • cells that produce the protein coded for by the target gene will have many mRNA molecules (floating around) that are complementary to the gene - easier to obtain • the mRNA is used as a template by reverse transcriptase to make cDNA (complementary DNA) from the mRNA template |
|
|
|
What is the principle behind producing DNA fragments using restriction endonuclease enzymes? |
• some sections of DNA have palindromic sequences of nucleotides (the base sequence on each strand reads the same in opposite directions) GAATTC -> reads: GAATTC CTTAAG <- reads: GAATTC • Palindromic sequences that are recognised by a restriction endonuclease are called ‘recognition sequences’ • Restriction endonucleases - enzymes that recognise recognition sequences and cut the DNA at these places e.g. EcoRI cuts at GAATTC, HindIII cuts at AAGCTT • if there are recognition sequences present at either side of the DNA fragment you want - use restriction endonucleases to separate the fragment from the rest of the DNA |
|
|
|
Describe how DNA fragments are produced using restriction endonuclease enzymes? |
1. DNA sample incubated with the specific restriction endonuclease (or 2 different endonucleases if the same recognition sequence isn’t present at either side of the fragment) 2. The enzyme(s) cuts the DNA fragment out (via hydrolysis reactions) NB - Sometimes a cut will leave a sticky end |
|
|
|
What is a sticky end? |
A small tail of unpaired bases at the end of a fragment left sometimes by a restriction endonuclease after DNA is cut |
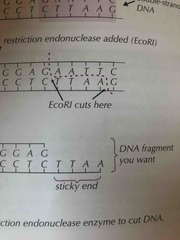
|
|
|
What is a sticky end? |
A small tail of unpaired bases at the end of a fragment left sometimes by a restriction endonuclease after DNA is cut (Click the hint to see) |
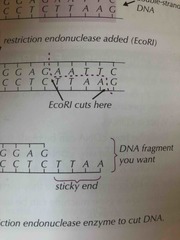
|
|
|
Why are sticky ends of a cut out DNA fragment useful? |
Can be used to bind the DNA fragment to another piece of DNA that has sticky ends with complementary base sequences |
|
|
|
How are the DNA fragments produced by using ‘reverse transcriptase’ and using ‘restriction endonuclease enzymes’ different? |
Using reverse transcriptase: • the DNA fragment only contains exons (complementary copy of mature mRNA) Using restriction endonuclease enzymes: • the fragments contain the whole base sequence of the gene, including the introns |
|
|
|
Why do different restriction endonucleases cut at different specific recognition sequences? |
The shape of the recognition sequence is complimentary to the enzymes active site (The recognition sequence is the substrate molecule) Different recognition sequences have different shapes. |
|
|
|
Why do different restriction endonucleases cut at different specific recognition sequences? |
The shape of the recognition sequence is complimentary to the enzymes active site (The recognition sequence is the substrate molecule) Different recognition sequences have different shapes. |
|
|
|
Why do different restriction endonucleases cut at different specific recognition sequences? |
The shape of the recognition sequence is complimentary to the enzymes active site (The recognition sequence is the substrate molecule) Different recognition sequences have different shapes. |
|
|
|
Here is a picture of restriction endonucleases at work! |

Back (Definition) |
|
|
|
What is the principle behind producing DNA fragments using a gene machine? |
Using technology: • Fragments of DNA can be synthesised from scratch, without the need for a pre-existing DNA or mRNA template (The DNA sequence doesn’t even have to exist naturally) • A database contains the necessary information to produce the DNA fragment - so any sequence can be made |
|
|
|
What is the principle behind producing DNA fragments using a gene machine? |
Using modern technology: • Fragments of DNA can be synthesis from scratch, without the need for a pre-existing DNA or mRNA template (The DNA sequence doesn’t even have to exist naturally) • A database contains the necessary information to produce the DNA fragment - so any sequence can be made |
|
|
|
In a gene machine, how can a DNA fragment be produced that contains a base sequence that doesn’t occur naturally? |
Sequences can be designed on computers and then stored in a database |
|
|
|
Describe how DNA fragments are produced using a gene machine? |
1. (If a base sequence doesn’t already exist) the sequence that is required is designed 2. The first nucleotide in the sequence is fixed to some sort of support (e.g. a bead) 3. Nucleotides added step by step in the correct order, in a cycle of processes that includes adding protecting groups NB - protecting groups make sure the nucleotides are joined at the right points, to prevent unwanted branching 4. This produces Oligonucleotides (short sections of DNA roughly 20 nucleotides long) |
|
|
|
Describe how DNA fragments are produced using a gene machine? |
1. (If a base sequence doesn’t already exist) the sequence that is required is designed 2. The first nucleotide in the sequence is fixed to some sort of support (e.g. a bead) 3. Nucleotides added step by step in the correct order, in a cycle of processes that includes adding protecting groups NB - protecting groups make sure the nucleotides are joined at the right points, to prevent unwanted branching 4. This produces Oligonucleotides (short sections of DNA roughly 20 nucleotides long) 5. Complete oligonucleotides are broken off from the support + all protecting groups removed 6. The oligonucleotides are joined together to make a longer DNA fragment |
|
|
|
What is the purpose of ‘protecting groups’ in a gene machine? |
Make sure the nucleotides are joined at the right points, to prevent unwanted branching of oligonucleotides |
|
|
|
What is the purpose of ‘protecting groups’ in a gene machine? |
Make sure the nucleotides are joined at the right points, to prevent unwanted branching of oligonucleotides |
|

