![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
99 Cards in this Set
- Front
- Back
|
Nucleotide |
Consists of a phosphate joined to a sugar (2'-deoxyribose) and a base |
|
|
Nucleoside |
Consists of a sugar (2'-deoxyribose) and a base |
|
|
Nucleobase |
Consist of purines: guanine (DNA and RNA), adenine (DNA and RNA). Also of pyrimidine: cytosine (DNA and RNA),thymine (DNA). |
|
|
Phosphodiester bond |
Nucleotides are joined by the 3'-hydroxyl of the 2'-deoxyribose sugar and by the 5'-hydroxyl. Create the sugar-phosphate backbone. |
|
|
5' and 3' ends |
DNA sequences occur from the 5'-phosphate end (on the left) to the 3'-hydroxyl end. The two strands of the double helix are wound around each other in an antiparallel orientation. |
|
|
Tautomeric states |
Formed by migration of ahydrogen atom, accompanied by a switch of asingle bond and neighboring double bond. The N- atoms attached to C , G and A are in the Amino- (NH2) --> imino (=NH) The O- atom in G and T are in Keto form Keto (C=O)--> Enol (C-OH) |
|
|
Base Stacking |
Bases are flat and water-insoluble (hydrophobic), which allows them to interact with each other through transient, induced dipoles (van der Waals) to increase DNA stability and lowers the free energy. |
|
|
Right-Handed Double Helix |
Polynucleotide chains in the double helix have their 5'-end facing toward the right. |
|
|
Minor Groove |
The angle at which the two sugars protrude from the base pairs is 120 deg. |
|
|
Major Groove |
The larger angle at which the two sugars protrude from the base pairs, 240 deg. Rich in chemical information. |
|
|
1953 |
Watson and Crick discovered the DNA structure. |
|
|
34 A/ 10.5 bp |
Measurement of one turn of the helix |
|
|
20 A |
Width of the double helix |
|
|
A conformation |
Observed at low humidity, 11 bp per turn, and compact major and minor grooves. |
|
|
B conformation |
Observed at high humidity, average structure of DNA, 10 bp per turn, with a wide major groove and a narrow minor groove. |
|
|
Z conformation |
Rare, stretched with 7-8 bps per turn, left-handed due to purines with syn confirmation. |
|
|
Denaturation |
When the complementary strands of DNA can be made to come apart at high temps (>100 C) or high pH. Typically reversible for DNA. |
|
|
Hybridization |
The ability to form hybrids between two single-stranded nucleic acids. Ex: 2 different types of DNA or DNA+RNA. |
|
|
abs. 260 nm |
The max amount of ultraviolet light DNA can absorb before breaking down (denaturing). |
|
|
Tm Melting Point |
The melting point of DNA at which point causes it to transition into a less ordered individual strand. |
|
|
Topology |
The ability to freely rotate to accommodate changes in the number of times the two chains of the double helix twist about each other. |
|
|
Superhelical density |
Denoted as σ, that represents the number of turns added or removed relative to the total number of turns in the relaxed molecule/plasmid, indicating the level of supercoiling. Commonly found as negatively supercoiling to help with separation of the two strands. |
|
|
Topoisomerase |
Enzymes that interrupt the sugar-phosphate backbone by introducing transient single or double stranded breaks into the DNA to change the linking number constraint. |
|
|
Histones |
Highly alkaline proteins found in eukaryotic cell nuclei that package and order the DNA into structural units called nucleosomes. They are the chief protein components of chromatin, acting as spools around which DNA winds, and playing a role in gene regulation. Without histones, the unwound DNA in chromosomes would be very long.
|
|
|
DNA gyrase |
Special topoisomerase II enzyme that introduces negative supercoils. |
|
|
Catenated |
Topoisomerases can link together circular DNA like two rings of a chain. |
|
|
1.7 |
turns per histone |
|
|
2' OH |
How RNA (sugar) differs from DNA (sugar). Hydroxyl group replaces the H atom in the backbone. |
|
|
Uracil |
RNA's unique nucleobase; differs from thymine by the absence of a 5-methyl group. |
|
|
Single Stranded |
Typically RNA is found as a ____ polynucleotide chain. |
|
|
Tertiary Structures |
RNA chains frequently fold back on themselves to form base-paired segments between short stretches of complementary sequences; similar to A form of DNA. |
|
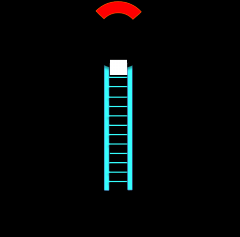
Stem Loop Structure |
The intervening RNA is looped out from the end of the double helical segment. |
|
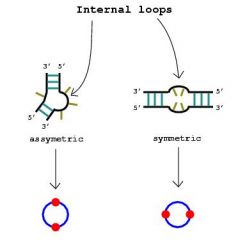
Internal Loop |
Unpaired nucleotides on either side of the stem |
|
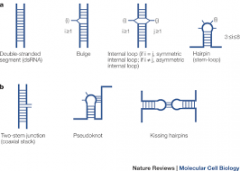
Bulges |
An unpaired nucleotide on one side of the buldge |
|
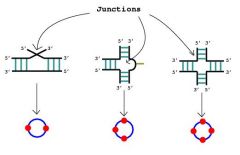
Junction |
Structural elements that form when three or more helices come together in space in the tertiary structures of RNA molecules
|
|
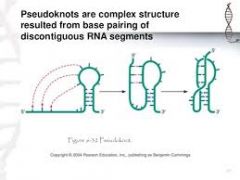
pseudoknot |
When base pairing occurs between noncontiguous sequences. |
|
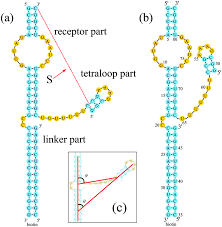
Tetraloop |
A type of four-base hairpin loop motifs in RNA secondary structure that cap many double helices; base-stacking interactions promote and stabilize it. |
|
|
non Watson-Crick base pair |
A feature of RNA that adds to its propensity to form double-helical structures. EX) G:U and G:A. |
|
|
Base triple |
RNA's flexible and rotational backbone allows for unusual hydrogen pairing between bases. EX) U:A:U |
|
|
Riboswitch |
Regulatory RNA elements that bind and respond to small molecule ligands in controlling gene transcription and translation. |
|
|
Nuclear Magnetic Resonance (NMR) |
High-resolution nucleic acid structure determination, in that the molecules are being observed in their natural solution; useful for molecules of up to 100 nucleotides. |
|
|
X-ray crystallography |
A tool used for identifying the atomic and molecular structure of a crystal, in which the crystalline atoms cause a beam of incident X-rays to diffract into many specific directions. By measuring the angles and intensities of these diffracted beams, a crystallographer can produce a three-dimensional picture of the density of electrons within the crystal.
|
|
|
Probing |
Biochemical techniques are used to determine nucleic acid structure, typically stems or loops.
|
|
|
Systematic evolution of ligands by exponential enrichment (SELEX) |
Generates novel RNA by synthesizing RNA molecules with randomized sequences to obtain sequence diversity. |
|
|
Aptamers |
RNA molecules selected biochemically that have an affinity for a specific small molecule or protein. |
|
|
Ribozyme |
An RNA that can also be an enzyme. Exhibit features of an enzyme: has an active site, a binding site for a substrate, and a binding site for a cofactor. |
|
|
RNase P |
One of the first ribozymes to be discovered that is involved in generating tRNA molecules from larger, precursor RNAs. |
|
|
1982 |
Nobel prize for discovering the structure of a ribozyme. |
|
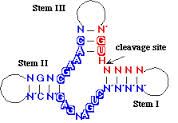
Hammerhead |
Ribozyme that cleaves RNA by the formation of a 2', 3' Cyclic Phosphate found in viroids; consists of three base-paired stems with a core of noncomplementary nucleotides required for cleavage. |
|
|
Viroids |
Infectious RNA agents of plants which depend on self cleavage to propagate. |
|
|
peptidyl transferase |
Enzyme that is responsible for peptide-bond formation during protein synthesis. |
|
|
Amino Group |
One of the functional groups of an amino acid that contains an NH2. pKa of about 9 |
|
|
Carboxyl Group |
One of the functional groups of an amino acid that contains a COOH. pKa of about 2 |
|
|
R-group (side chain) |
One of the functional groups of an amino acid that has 22 variations. Determines the specific characteristics of an amino acid. |
|
|
L conformation |
Every amino acid (except glycine) can occur in two isomeric forms, because of the possibility of forming two different enantiomers (stereoisomers) around the central carbon atom. Only type of amino acids that are manufactured in cells and incorporated into proteins. |
|
|
pKa |
The acid dissociation constant at logarithmic scale that measures weak acids. |
|
|
Peptide Bond |
The covalent links between amino acids in a protein. |
|
|
Glycine |
The smallest amino acid which has hydrogen as its side chain. It is a neutral-nonpolar, hydrophobic, colorless, sweet-tasting crystalline solid. It is achiral and extremely flexible. It can fit into hydrophilic or hydrophobic environments |
|
|
Proline |
The only amino acid with a secondary amine side chain. The exceptional conformational rigidity affects the secondary structure of proteins by forming a double bond. Neutral-nonpolar and hydrophobic. |
|
|
Cysteine |
Neutral-polar and hydrophilic and forms Disulfide bond. The sulfhydryl (-SH) on its side chain makes it sensitive to oxidation-reduction reactions. |
|
|
Disulfide Bond |
A single covalent bond between the sulfur atoms of two amino acids called cysteine |
|
|
Extracellular protein |
Proteins on the cell surface and in extracellular space are exposed to an environment with a redox potential that favors disulfide bond formation, which enhance its stability of a folded protein by adding covalent cross-links. |
|
|
Water |
A hydrogen bonded liquid that can donate two hydrogen bonds and accept two hydrogen bonds. |
|
|
Hydrophilic |
Molecules that participate in the network of hydrogen bonds of water. Disperses into an aqueous solution; soluble. |
|
|
Hydrophobic |
Molecules that perturb the network of hydrogen bonds of water. Insoluble molecules that remain adjacent to each other. |
|
|
Primary structure |
The sequence of amino acid residues in the polypeptide chain. One-dimensional string, specifying a pattern of chemical bonds. |
|
|
Secondary structure |
The local conformation of a protein's polypeptide chain, 3D arrangement of a short stretch of amino acid residues. Contains two types of conformations: alpha helix and beta strand. |
|
|
Alpha helix |
The polypeptide backbone spirals in a right handed sense around the helical axis, so that hydrogen bonds form between the main-chain carbonyl group of one residue and the main-chain amide group of a residue four positions further along in the chain. |
|
|
Beta Strand |
An extended conformation in which the side chains project alternately to either side of the backbone, and the amide and carbonyl groups project laterally. The backbone is slightly pleated. |
|
|
Tertiary structure |
The usually compact, 3D folded arrangement that an entire polypeptide chain (single domain or multidomain) adopts under physiological conditions. |
|
|
Quaternary structure |
When individual, folded polypeptide chains associate with each other to form multiple protein subunits in a larger complex. |
|
|
Structural domain |
A part of a polypeptide chain with a folded structure that does not depend for its stability on any of the remaining parts of the protein. Typical proteins are b/w 50-300 amino acid residues so this helps with the longer proteins to reduce errors. |
|
|
Ribbon diagram |
A representation that emphasizes the role of secondary structural elements in the folded conformation of a domain. Alpha helices are curled ribbons and beta sheets are curved arrows pointing toward the carboxyl terminus. |
|
|
Post-translational modification |
Modifications of amino acid side chains, introduced following emergence of the polypeptide chain from a ribosome, can modulate the structure and function of a protein. |
|
|
Glycosylation |
Modification of an amino acid through the addition of one or more sugars (glycans) to an asparagine side chain or to a serine or threonine side chain. Characteristic of ectodomains of cell-surface proteins and of secreted proteins. |
|
|
Phosphorylation |
Serine, threonine, tyrosine, or histidine side chain modifications that adds a phosphate group. |
|
|
Denaturation |
When a protein is exposed to high concentration of solutes and begins to unfold into a random coil. |
|
|
Renaturation |
When a protein is removed from high concentration of solutes allows the protein to refold. |
|
|
Folding Chaperones |
Unfold a misfolded protein and allows it to try again. Aids during renaturation. |
|
|
Chromosome |
DNA molecule and its associated protein. It is a compact form of the DNA that readily fits inside the cell. Serves to protect the DNA from damage by making it extremely stable. Allows for easier transmission during cell division. Organizes each molecule of DNA and regulates its accessibility. |
|
|
Linear DNA |
Eukaryotic cells have multiple ____ chromosomes. |
|
|
Circular DNA |
Prokaryotic cells mostly have this singular type of chromosome. Requires topoisomerases to separate the daughter molecules after replication. |
|
|
Histones |
The associated proteins found on a given region of DNA (chromatin). Small, positively charged proteins. Five in total within eukaryotic cells, four are core proteins. |
|
|
Escherichia coli |
Number of Chromosomes: 1 Chromosome Copy Number: 1 Forms of Chromosome: Circular Genome Size (Mb): 4.6 |
|
|
Saccharomyces cervisiae |
Number of Chromosomes: 16 Chromosome Copy Number: 1 or 2 Forms of Chromosome: Linear Genome Size (Mb): 12.1 |
|
|
Homo sapiens |
Number of Chromosomes: 22 + X & Y Chromosome Copy Number: 2 Forms of Chromosome: Linear Genome Size (Mb): 3200 |
|
|
Plasmid |
Prokayotes typically one or more smaller independent circular DNAs that are not essential for bacterial growth but contain desirable traits, such as antibiotic resistance. |
|
|
Diploid |
Eukaryotic cells contain two copies of each chromosomes. One from each parent, unless they are homologus sex cells. |
|
|
Polyploid |
Some organisms have more than two copies of each chromosomes |
|
|
Gene density |
The average number of genes per megabase of genomic DNA. |
|
|
microsatellite DNA |
Composed of very short (<13bp) tandemly repeated sequences of DNA. The most common being dinucleotide repeats (CACACACA). This arises from difficulties in accurately duplicating the DNA. |
|
|
Genome-wide repeats |
Larger tandemly repeated sequences of DNA, typically >100bp. These sequences can move from one place in the genome to another (transposable). |
|
|
Junk DNA |
Repeated DNA; can either be genome-wide repeats or microsatellite DNA |
|
|
Core Histones |
8 histones with two of each type. H2A, H2B, H3, & H4. Present in equal amounts throughout the cell. |
|
|
nucleosome |
Composed of a core of eight histone proteins and the DNA wrapped around them. |
|
|
Linker histone H1 |
Histone that binds to the DNA between each nucleosome that is between 20-60 bp long. |
|
|
Histone tail |
The core histone has an amino-terminal extension that lacks a defined structure. Proteases like trypsin can cleave it. posttranslational modifications occur here that alter the function of individual nucleosomes. |
|
|
30 nm fiber |
Nucleosomal DNA can form more complex structures (after H1 has been added) in high salt concentrations |
|
|
Chromosome scaffold |
DNA can be folded even further after 30 nm fiber. The 30 nm fiber forms loops of 40-90 kb that are held together at their bases. |

