![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
118 Cards in this Set
- Front
- Back
|
In Prokaryotic DNA replication, this enzyme binds the sequence at the ori C site containing 11 GATC/CTAG repeats |
DnaA |
|
|
In Prokaryotic DNA replication, this enzyme methylates Adenine sites |
Dam methylase |
|
|
Helicase enzyme in prokaryotic DNA replication |
DnaB |
|
|
Topoisomerase enzyme in prokaryotic DNA replication |
DNA gyrase (target for drugs) |
|
|
Major 'worker' of replication in prokaryotic DNA replication; responsible for building the new DNA strand |
Polymerase III |
|
|
Primase enzyme in prokaryotic DNA replication |
DnaG |
|
|
In prokaryotic DNA replication, these enzymes remove RNA primer and replace sequence with DNA |
Rnase H and DNA polymerase I |
|
|
What is type 2 topoisomerase used for in prokaryotic DNA replication? |
Unlink newly synthesized circular chromosomes allowing cell division to continue |
|
|
This enzyme binds the DNA sequence at the points of origin in Eukaryotic DNA replication |
Origin recognition complex (ORC) |
|
|
This enzyme binds DNA and ORC in Eukaryotic DNA replication |
Minichromosome maintenance proteins (MCM) |
|
|
What are SSBs named in Eukaryotic DNA replication? |
RPA |
|
|
In Eukaryotic DNA replication this wraps around the DNA and travels along the strand with DNA polymerase |
PCNA (sliding clamp) |
|
|
In Eukaryotic DNA replication this works with Primase to create 10 nucleotide RNA primer followed by 20 nucleotides |
Pol a |
|
|
What enzyme, in Eukaryotic DNA replication, continues DNA strand elongation on the leading strand? |
Pol e |
|
|
What enzyme, in Eukaryotic DNA replication, continues adding dNTPs to the Okazaki fragment on the lagging strand? |
Pol o |
|
|
In Eukaryotic DNA replication what is primer removed by? |
Flag endonuclease 1 (FEN1) and RNase H |
|
|
In Eukaryotic DNA replication what fills in missing DNA sequence? |
DNA polymerase o |
|
|
What is the purpose of Telomerase in Eukaryotic DNA replication? |
Lengthen 3' overhang by using built in primer that recognizes the TTAGGG sequence; Telomerase acts as a polymerase increasing the length of parental strand; allows primes to bind and new strand to lengthen |
|
|
What is the purpose of 3'-5' exonuclease? |
Excise mismatched nucleotide |
|
|
Missense mutation |
Changes a letter that changes the amino acid sequence |
|
|
Silent mutation |
Missense mutation that does not change the amino acid sequence |
|
|
Nonsense mutation |
Changing letter that makes sequence stop |
|
|
What gene mutation can cause disease? |
Triplet expansion |
|
|
What does UV radiation cause in DNA? |
Thymine dimers/cyclobutane ring |
|
|
What mechanism would be most efficient to correct deprivation? |
Base excision repair |
|
|
Core enzyme of prokaryotic polymerase |
5 subunits |
|
|
Holoenzyme of prokaryotic polymerase |
core enzyme + sigma (o) factor |
|
|
Process of Rifampin |
Inhibits transcription by binding to the B subunit of the RNA polymerase (stops phosphordister bond formation in the RNA) |
|
|
What is Rifampin used to treat? |
Rhodococcus, Mycobacteria, and Staphylococci |
|
|
In Eukaryotic Transcription, what is RNA polymerase II responsible for? |
protein-coding genes |
|
|
In Eukaryotic transcription, what is always used by RNA polymerase II to initiate transcription? |
General transcription factors |
|
|
In Eukaryotic transcription, what are the proteins involved for recruitment of general transcription factors and RNA polymerase to the promoter |
Regulatory transcription factors |
|
|
What is the mechanism of amanitin? |
Binds to RNA polymerase II and inhibits transcription |
|
|
What symptoms are exhibited after ingestion of Amanita phalloides? |
Gastrointestinal (vomiting, diarrhea, and cramps), recovery period prior to kidney and liver failure, eventually death |
|
|
What inhibits eukaryotic RNA polymerase II? |
a-amanitin |
|
|
What inhibits all of RNA polymerases? |
Actinomycin D |
|
|
What is Actinomycin D's mechanism of action? |
Stabalizes topoisomerase-I on the DNA and prevents RNA polymerase progression, also causing double-strand breaks |
|
|
What RNA polymerase is used as a chemotherapy drug for lymphomas and various sarcomas? |
Prevents transcription in both eukaryotes and prokaryotes |
|
|
What is inclusion or exclusion of a sequence in splicing lead to ? |
mutations; will interfere with production of the correct protein |
|
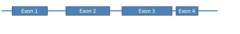
Your patient has a novel mutation disrupting the splice donor site of intron 2. Which of the following mRNAs is likely a result form this mutation? |
|
|
|
What are protein isoforms? |
variations of a protein with slightly different functions (alternative splicing) |
|
|
What does alternative splicing allow? |
Allows the cell to tailor the protein for the desired job in that cell type |
|
|
What is alternative splicing? |
using different combinations of splice sites allowing for the production of different isoforms form the same gene |
|
|
Intron retention |
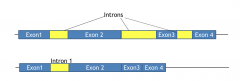
mRNA contains and will encode protein the sequence of this intron |
|
|
Exon skipping |
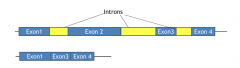
mRNA lacks and will not encode protein front the sequence of exon2 |
|
|
Mutually exclusive exons |
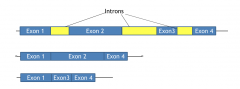
Will allow either exon 2 or exon 3 to be included in the mRNA, but not both |
|
|
Alternative polyadenylation |
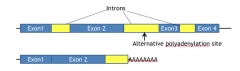
mRNA contains and will encode protein from the sequence from intron 2 |
|
|
Proteolysis |
Cleavage and removal of amino acid residues in the protein |
|
|
Glycosylation |
Addition of oligosaccharides to proteins via a glycosidic bond to the hydroxyl or amine group of an amino acid side chain |
|
|
Methylation and acetylation |
addition and removal of methyl and acetyl groups at specific lysine and arginine residues controls gene expression |
|
|
Myristolylation |
myristoyl (C14) is added to the N-terminus at glycine residue (must cleave start methionine; results in protein association with intracellular vesicles) |
|
|
Palmitoylation and prenylation |
addition of a lipid on a cystein side chain |
|
|
Hydroxylation |
addition of hydroxyl group to amino acid (proline and lysine residues are hydroxylated on the a-chians of collagen) |
|
|
Carboxylation |
carboxyl groups are added to glutamate residues by a vitamin K dependent carboxylation (important for blood clotting factors |
|
|
Ubiquitination |
addition of small polypeptide ubiquitin to a protein via a lysine residue to target proteins for degradation in the proteasome |
|
|
Gated transport |
the nuclear pore complex facilitates protein transport from cytosol to nucleus |
|
|
Transmembrane transport |
membrane-bound protein translocators directly transport specific proteins across the membrane (proteins usually unfolded to undergo this transport) |
|
|
Vesicle transport |
membrane enclosed transport intermediates pinch off from other membrane-enclosed compartments |
|
|
Signal sequence |
short sequence in the protein that is used to direct proteins to the correct compartment of the cell; sequence can be cleaved from protein once transport has occurred |
|
|
Signal patches |
3-D arrangement of amino acids on the proteins surface that forms after the protein is folded; can be used for gated-transport |
|
|
Sorting receptors |
recognize corresponding signal sequences or patches and guide protein to the correct location |
|
|
What types of proteins are transported into the nucleus? |
Histones, transcription factors, polymerases |
|
|
Nuclear localization signal (NLS) |
signal sequence or signal patch that is rich is positively charged amino acids |
|
|
Nuclear import receptors |
soluble cystosolic proteins that bind the NLS as well as nucleoporins |
|
|
TOM complex |
translocase for the outer membrane |
|
|
TIM complex |
translocases for the inner membrane (TIM23 and TIM22) |
|
|
OXA |
translocase for inner membrane |
|
|
Proteins transported into ER |
transmembrane proteins, proteins found int he lumen of the ER or other membrane-bound organelles, and excreted proteins |
|
|
Free ribosomes |
remain in the cytoplasm synthesize proteins used in the cytoplasm |
|
|
Membrane-bound ribosomes |
start in the cytoplasm and are recruited to the surface of the ER during translation |
|
|
ER sequence |
contains a section of hydrophobic/nonpolar amino acid |
|
|
Signal-recognition particle (SRP) |
bind the ER signal sequence, temporally stalls translation |
|
|
Signal-recognition particle receptor (SRP-receptor) |
an integral membrane receptor on the cytosol side of the ER membrane. Bring SRP-ribosomes to the surface of the ER |
|
|
Sec61 complex |
ER protein translocator, donut-shaped, contains binding site for the ER signal sequence |
|
|
Processes within the Golgi |
Glycosylation, sulfation, phosphorylation, proteolsis |
|
|
Proteins that bind the cytosol side of the vesicles used to transport proteins to/from/within the Golgi |
Clathrin, COP I, COP II |
|
|
In protein transport to the lysosome, what targets the protein for the lysosome? |
the attachment of mannose 6-phosphate to a protein |
|
|
A deficiency of the ability to phosphorylate mannose leads to what? |
I-cell disease (mucolipidosis II) |
|
|
Symptoms of mucolidosis II |
skeletal abnormalities, restricted joint movement, coarse facial features, severe psychomotor impairment, death usually occurs by 8 years of age |
|
|
Symptoms of mucolidosis II in cats |
abnormal facial features, retarded growth, progressive hindlimb paresis, skeletal abnormalities (major thing) (mutation in gene encoding GIcNAc-phophotranserase) |
|
|
Constitutive secretion |
secretory vesicle travels from Golgi directly to the membrane |
|
|
Regulated secretion |
secretory vesicles remain in the cytoplasm util a signal triggers the fusion and release of contents |
|
|
What do clathrin-coated vesicles transport? |
macromolecules bound to specific receptors for import into the cell |
|
|
How is cholesterol imported into the cell? |
bound to low-density lipoproteins (LDL) by binding the LDL receptor (LDLR) and being transported to the lysosome via vsicles |
|
|
What is needed for translation? |
ribosomes (multiple proteins and rRNAs), aminoacyl tRNA (transport amino acids), mRNA (transported to the cytoplasm), translation factors (facilitate the process at each step) |
|
|
Shine Dalgarno sequence |
ribosome binding site ~10 bases upstream from the start codon, 5' AGGAGGU 3', base pairs with the 16S rRNA (30S) |
|
|
IF-1: initiation factor 1 |
binds 30S subunit, prevents subunits joining prematurely |
|
|
IF-2: initiation factor 2 |
binds initiator-tRNA, hydrolyzes GTP |
|
|
IF-3: initiation factor 3 |
stabilizes free 30S subunit, prevents subunits from joining prematurely |
|
|
EF-Tu in elongation |
escorts aminocyl-tRNA to the ribosome and checks for correct codon-anticodon base pairing. hydrolyzes GTP |
|
|
Ribosomes in elongation |
catalyzes peptide bond formation through 23S rRNA acting as a ribozyme |
|
|
EF-G in elongation |
binds ribosomes to facilitate translocation. Hydrolyzes GTP |
|
|
Eukaryotic translation initiation factors |
Ternary complex, recruitment complex, eIF3, eIF5B |
|
|
eIF2 |
binds the initiator tRNA (mehtionine) and GTP |
|
|
eIF4E |
binds 5' cap |
|
|
eIF4G |
binds eIF4E and poly-A binding protines (PABP) |
|
|
eIF4A |
helicase that unwinds seoncdary structure in the 5' UTR |
|
|
eIF3 |
binds the 40S subunit, brings subunit to the pre-initiation complex |
|
|
eIF5B |
binds the 60S subunit, brings subunit to the initiation complex |
|
|
Ternary complex |
eIF2, GTP, aminoacyl-tRNA |
|
|
Recruitment complex |
eIF4E binds 5' cap of mRNA, eIF4G, POBP, eIF4A |
|
|
Enzymes in elongation |
eEF-1, ribosome, eEF-2 |
|
|
eEF-1 in eukaryotic elongation |
escorts aminoacyl-tRNA to the ribosome and checks for correct codon-anticodon base pairing. Hydrolyzes GTP |
|
|
Ribosomes in eukaryotic elongation |
catalyzes peptide bond formation through 28S rRNA acting as a ribozyme |
|
|
eEF-2 in eukaryotic elongation |
binds ribosomes to facilitate translocation. Hydrolyzes GTP |
|
|
What is the mechanism of antibiotics? |
Inhibitors of prokaryotic translation and bind directly to the ribosome |
|
|
What is the mechanism of chemotherapeutics? |
eukaryotic translation inhibitors since translation is essential for cell survival and occurs at a higher rate in rapidly proliferating cells ex: targets for eIF4E, eIF4A |
|
|
Scientists are designing a new chemotherapeutic drug that will target translation initiation. What is a protein that is a likely target? |
eIF4E |
|
|
Transcriptional activators (genetic switches) |
turns genes on; increases transciriton by enhancing the binding of the polymerase to the promoter or opening the helix |
|
|
Transcriptional repressors (genetic switches) |
turns genes off; binds the operator, a sequence upstream of transcription, blocks access to promoter for RNA polymerase
|
|
|
What are Lac operon and Trp operon? |
genetic switches |
|
|
Eukaryotic regulation of transription |
promote/prevents assembly of transcription initiation complex (modification of chromatin, DNA methylation, gene-spcific transcription factors) |
|
|
Purpose of chromatin modifications? |
transcription factors cannot bind the promoter if DNA is tightly wrapped n nucleosomes |
|
|
Mechanisms of chromatin modifications |
covalent histone modifications, nucleosome removal, nucleosome replacement, nucleosome remodeling |
|
|
Global regulation |
changes in the level of all protein synthesis in the cell |
|
|
Specific regulation |
changes in the translation level of specific mRNAs in the cell |
|
|
How do cells decrease protein synthesis in times of high stress? |
phosphorylation of eIF2 |
|
|
What is the mechanism of ricin? |
binds and removes an adenine residue from 28S rRNA making the ribosome inactive |

