![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
44 Cards in this Set
- Front
- Back
|
What are enzymes? |
proteins however there can be some RNA enzymes. |
|
|
what is the function of an enzyme? |
they are biological cataylsts, that lower the activation energy making it easier for the transition state to be reached so that a reaction can take place more rapidly. they DO NOT alter the equilibrium and do not alter the ratio of production to reactant. |
|
|
what are the four features of an enzyme catalysed reaction? |
-much faster. -milder reaction conditions (are efficient at mild conditions of temp and pH inside cells). -very specific to substrate. -tightly regulated (ensuring activity is efficient when and where its needed). |
|
|
What is the active site of an enzyme and what is its function? |
is a 3D structure where the substrate will bind (initially with non-covalent bonds) and is where the reaction will take place. -determines the specificity of enzyme. |
|
|
what is the role of the amino acids leading into the active site |
these will interact with the substrate, some will hold it in place whilst others will be involved with overall reaction. |
|
|
Describe the two types of enzyme specificity. |
-Geometric: substrate must be correct shape to fit into active site. -Stereospecificty: the substrate must be of the correct orientation to be able to bind to the active site. since enzymes have asymmetric binding sites.
substrate must have both of theses to be able to bind to an enzyme.
|
|
|
what are the steps of an enzyme catalysed reaction? |
1. formation of enzyme substrate complex 2.transition state. 3.formation of products. |
|
|
what are the two models of enzyme-substrate interactions? |
-lock and key: there must be an exact match of substrate to enzyme. -induced fit model: initially there is not a perfect fit between the two, once bound they will induce a change to form a perfect fit and then a product will be formed. this is a dynamic relationship. |
|
|
describe the catalytic mechanism of 'Covalent Catalysis' |
involves the formation of a covalently bound enzyme-substrate intermediate which is highly reactive, so will easily enter the transition state and complete the reaction. EG: Serine Protease enzymes such as trypsin and chymotrypsin. |
|
|
Describe the catalytic mechanism of 'Acid- Base Catalysis' |
involves the donation/ acceptance of a proton and is very common. EG; Histinde is particularly good since it has a pka of 6.5 so depending on pH environment it will either accept or donate protons. other commonly involved sidechains are Glu, Asp,Lys, Arg, and Cys. |
|
|
what are the two types of co-factors and what do they do? |
metal ions and co-enzymes . both of which work to help enzyme function.
|
|
|
what is the name for a complete functional enzyme and its cofactor? |
Holoenzyme.
|
|
|
what are most coenzymes derived from and what are they? |
-they are small organic molecules derived form vitamins. hence if you have a vitamin deficiency, it can effect enzyme function causing a defect in cells ability to operate properly.
|
|
|
how do you name enzymes? |
x.y.z.[a
x=class y=sub class z=sub sub class a=serial number, not all enzymes have this. |
|
|
what are the six enzyme classes? |
1. Oxidoreductase (transfer electrons). 2. Transferase (group transfer reactions). 3. Hydrolase (h20 transfer). 4. Lyases (+/- of double bonds). 5. Isomerase (transfer to yeild isomeric forms). 6. Ligase (condesation reactions coupled to ATP cleavage). |
|
|
what are enzyme reactions usually characterised by? what is the rate dependent upon and what dose the graph look like? |
-the initial velocity, (this is a linear line to the hyperbola). -dependent on the availability of substrate, as more is converted to product rate will slow down (hyperbola on graph). -time on x-axis, conc. of product on y-axis. |
|
|
what effect dose enzyme concentration have when substrate is in excess? |
has a proportional linear relationship, as more enzyme is added the rate of reaction will increase. |
|
|
Describe what is occurring on an enzyme substrate graph? |
-initially at low [s], V increase linearly .. is a first order reaction. -as [s] increases the rate increases, but will gradually drop off as more enzyme active sites a occupied. -eventually it will reach a point where the enzyme is saturated and Vmax. at this point the reaction is independent of the [s] so is a zero-order reaction. |
|
|
what is the Michaelis Menten equation? |
Vmax [S] V= KM + [S]
|
|
|
What are the two enzyme parameters and what do they allow for? |
enzyme comparisons. -Vmax= the max velocity of reaction, when enzyme is saturated. - Km= is the [s] that gives you half of the Vmax velocity. |
|
|
what are the assumptions made for the Michaelis-Menten equation |
-rate of formation= rate of breakdown, and rapidly will reach a steady state. -[s]>> [E] -[s] does not change during initial stages of reaction. -product is in such small amounts for initial stages of reaction that it can be ignored. |
|
|
how many ways can ES be broken down and how many forms will E exist in at any point of reaction? |
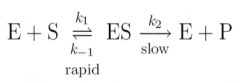
|
|
|
what is the Km of a substrate enzyme pair relative two and what is the unit of Km? |
relative to there affinity, so can be essentially thought of as dissociation constant for binding/release of enzyme. unit = mmol per L |
|
|
what does a low Km tell you |
that there is a high binding affinity between enzyme and substrate. |
|
|
what does a lineweaver-burk plot look like and what does it tell you? |
its a linear graph of 1/V vs. 1/[s] -where the line intercepts the y-axis it will tell you the value of 1/Vmax. -where is intercepts the x-axis it gives you a values of -1/Km |
|
|
what is Kcat? |
This is the turnover number (the num of moles of substrate converted to product per mole of enzyme per second). Kcat= Vmax/[Et] |
|
|
what does the ratio of Kcat/Km tell you? |
its a measure of the enzymes efficiency (describes how quickly bound s can be converted to P relative to how easily S is bound to E). -High ratio indicates efficient enzyme !
|
|
|
what is the advantage of phsyiological [S] being the same or lower than Km? |
since enzyme is only working at half Vmax it means that there is possibilities to up/down regulate activity, allowing increased cells ability to respond to changing conditions. |
|
|
what is an enzyme inhibitor and why are they helpful? |
its a compound that binds to an enzyme and reduces its activity. this is important for natural metabolism regulation, also many inhibitors are drugs, toxins and poisons. -can be helpful to study enzyme mechanisms and metabolic pathways. |
|
|
what are irreversible enzyme inhibitors? |
these bind covalently to enzymes and inactivate them irreversibly, most of these will be natural toxins. -they get into active site and form covalent bonds and get 'stuck' there, blocking the active site hence down regulating cell production. EG: Penicillin, inhibits glycopeptide transpeptidase found in bacteria cell wall. |
|
|
what are Competitive inhibitors? |
these are a type of reversible inhibitor that will bind/release in a concentration dependent manner. -they bind at the active site, preventing substrate from binding. -inhibition can be overcome by adding more substrate, so the Km value will increase (since its harder for substrate to bind). but the Vmax will remain the same, since its still possible to saturate enzymes... its just harder. |
|
|
what are non-competitive inhibitors? |
these are a type of reversible inhibitor that will bind/release in a concentration dependent manner. -they do not bind to the active site, but binding causes a conformational change that will mean enzyme does not work when its bound. -hence the Km will not be altered but the ability to reach Vmax is effected so the value of Vmax will be reduced. |
|
|
what are allosteric enzymes? |
have a conformational change on binding of a modulator and often have several protein subunits, and binding sites are not located at the active site. -follow a sigmoidal curve, indicating cooperative behaviour.
|
|
|
what effects will an allosteric activator and inhibitor have on an allosteric enzyme? |
an activator will shift the curve to the left (increasing Vmax), whilst and inhibitor will shift the curve to the right (lowering Vmax). |
|
|
how many allosteric regulators can bind to an allosteric enzyme at one time? |
multiple!
|
|
|
allosteric enzymes are often involved in what sort of system? |
feedback! where the product for the first reaction with be the substrate for the following reaction and so on. sometimes the final product will then work back to inhibit what ever was activating the first step. |
|
|
what are control proteins? |
these can stimulate or inhibit some enzymes with the intention of affecting their activity. |
|
|
what enzymes can be activated by proteolytic cleavage? |
Zymogens! |
|
|
covalent modification is another means of.... |
regulating enzyme activity. EG: phosphorylation |
|
|
what are isoenzymes and how can they be used? |
they are often multi-subnit. and will have the same function, but are coded for by different gene sequences so will perform the same action in slightly different manners depending on the tissue they are found in. this enables them to act as tissue markers, by each tissue displaying a tissue specific pattern. |
|
|
how can enzymes be used as drug targets and what is an example of this? |
A --e1--> B --e2--> C -if over production of B causes damage, then can target e1 to reduce B production. -or if not enough B then target e2 so that then B is produced but not used up to make C. EG: gout, target enzyme to reduce uric acid production |
|
|
what are the modes of inhibition that can be used against HIV? |
both competitive and non-competitive inhibition. |
|
|
In what way can HIV competitive drug designs function? |
-the HIV reverse transcriptase can be inhibited by nucleoside analogs (compounds that look like the substrate of the enzyme). - or you can inhibit the function of HIV protease. |
|
|
How does non-competitive drug design against HIV function? |
non-nucleosides that will bind somewhere other than the active site on the enzyme |

