![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
110 Cards in this Set
- Front
- Back
|
Causes of DNA mutations |
Spontaniously Induced |
|
|
Spontaneous mutations |
Naturally occuring in all cells |
|
|
Spontaneous mutations sources |
Errors in DNA replication Spontaneous lesions |
|
|
Induced mutations |
From extrensic sources (mutagens ) |
|
|
How mutagens induced mutations |
Add base analoge Alter base -->mispairing Damage base |
|
|
Prevention of errors before they happen |
Anti ROSs mechanisms |
|
|
Direct reversal of Damage |
UV make Cyclobutane Pyrimidine Dimers . By O6-methylguanine methyltransferase it repair |
|
|
Excision Repair Pathways |
General Coupling of Specific Mismatch |
|
|
General excision repair |
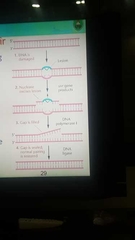
Nucleotide excision repair AKA |
|
|
Defect in Nucleotide excision repair lead to |
Xeroderma Pigmentousm |
|
|
XP Proteins |
XPA & XPG |
|
|
XP Proteins fx |
Damage recognition Enzyme activites( endonuclease and helicase ) |
|
|
Coupling of Transcription and repair |
RNA polymerase is paused when encountering a lesion |
|
|
Cockayne's Syndrome |
Mutation in CSB protein |
|
|
CSP protein fx |
Recognize that the RNA polymerase is stalled due to a mutation |
|
|
base excision repair |
single abnormal bases are recognized and removed |
|
|
What is DNA glycosylase |
enzyme that work on N-glycosidic bond bet. nitrogen base and deoxyribose of DNA backbone yields to apuritic or apyremidinc site (AP) |
|
|
abnormal bases that dna glycosylase works in it |
hypoxanthine from adenine bases damaged by oxidation or ionaizing radiation |
|
|
(AP) repaired by
|
AP endonuclease |
|
|
AP endonuclease mechanism of action |
cleaves PDE bond --------DOR is removed ------polymerase and ligase complete the mechanism |
|
|
Mismatch repair system in human |
hMSH 2 and 1 |
|
|
mismatch repair sys in bacteria |
Mut S and Mut L |
|
|
mismatch repair sys in bacteria recognition of mismatch |
adenine methylase mehylate the new DNA in delayed time so the delayed time is the target of MSH system in bacteria |
|
|
HNPCC and mismatch system in human |
7.5% of all cancer cases are caused by mutation in MSH and the most of other 7.5% are caused by mutation in MLH 15 ----* 50% |
|
|
Translesion DNA synthesis |
specialized DNA polymerases are capable of replicating across a site of DNA damage |
|
|
specialized DNA polymerases properites |
1.low fidelity when copyingundamaged DNA 2.lack 3' to 5' proofreading activity error rates range from 100 to 10,000 times higher than theerror rates of the normal replicative DNA polymerases 3. selectivity insertion lead to double strand breaks |
|
|
recombinational repair use |
double strand breaks in DNA |
|
|
recombinational repair pathways |
*Non-homologous end joining -----) high frequency of errors resulting from deletion of bases around the site of damage *Homologous repair with the undamaged chromosome iiiiiiiiiiiiis better |
|
|
key proteins in Homologous repair |
RecA Rad51 protein dna filaments |
|
|
BRCA 1& 2 role |
activate homologous recombination repair of DNA double-stranded breaks by recruiting Rad51 to the ssDNA. |
|
|
BRCA1 |
is also involved in transcription and transcription-coupled DNA repair. |
|
|
breast cancer and BRCA1&2 |
Mutations in BRCA1 account for 2% of all breast cancers and, at most, 5% of ovarian cancer. |
|
|
Processing of mRNA in EKs types |
Capping Splicing Polyadenylation |
|
|
Mechanism of capping |
After 25 nucleotides of RNA the 5' end of it modified by GTP in different orientation |
|
|
What is the type of bond between cap and modified RNA |
5' to 5' triphosphate bridge |
|
|
Importence of capping |
-Stabilizes the mRNA . -Differntiate the EK mRNA from other RNAs. -Cod the mRNA to be exported by CBC to the cytoplasm -In translation |
|
|
RNA splacing is catalyzed by |
Splicisomes |
|
|
Spliceosomes are composed of |
snRNA |
|
|
snRNA contents |
Proteins and snRNA |
|
|
When Polyadenylation happens |
After cleave of 3' end of mRNA that has a sequence of AAUAAA |
|
|
Enzyme that is responsiple of polyadenylation |
Poly-A-polymerase |
|
|
Precursor of ATP in polyadenylation is |
ATP |
|
|
Significance of polyadenylation is |
-help transportion of mRNA to cytoplasm -translation -stabilizes it |
|
|
Where the alternative polyadenylation taken place |
If the protein coding genes have more than one polyadenylation site |
|
|
Alternative polyadenylation produce |
3'-UTR (3' UNTRANSLATED REGION ) |
|
|
Example of alternative polyadenylation |
Calsitonin in thyroid and CGRP in brain are from the same gene |
|
|
MicroRNAs role |
Causing degradation or blokage of translation of mRNA |
|
|
How microRNAs are affect mRNA |
Bind to complementary sequences of mRNA |
|
|
Alternative polyadenylation and microRNA relationship |
More 3'-UTR from alternatinve polyadenylation would contain binding sites of MicroRNAs and lead to repress translation and degredation of mRNAs |
|
|
SNPs and alternative polyadenelation relationship |

SNPs may include AAUAAA sequence in polyadenelation signal that required for cleavage so interupte the adding of microRNA so more protein is producted |
|
|
Degradation of mRNA |
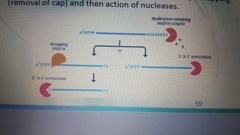
|
|
|
EK an bacterial difference in mRNA stability |
In EK the mRNA is more stable |
|
|
Gene amplification defention |
Multiplying the number of copies of a specific region of chromosome -)increase DNA in that region --) more mRNA from that region ---)more specific protein production |
|
|
Gene amplification and cancer cells |
1-all cancer cells use gene amplification to produce more DHF reductase to resiste the methotrexate 2-breast tumor cells amplify HER2 to be more sensitive to growth |
|
|
Alternative splicing in UDP glucuronsyltransferase |
Catalytic subunit of the enzyme is constant in all the body( 2.3.4.5 genes ) but the substrate binding subunit are variable (1 gene) because 5' region of the UGT1A complex contain 9 variables tandemly arrayed first axon tandomly arraayed |
|
|
General Transcription Factors fxs |
|
|
|
Core components of Promoter |
TATA Box binding site of TF11B BRE Upstream element Inr initiator element Multiple Downstream Elements |
|
|
Promoter Proximal Elements -1 |
Upstream of core promoter region |
|
|
Promoter Proximal Elements-2 |
Important for strong expression |
|
|
Promoter Proximal Elements can be |
Gene specific and bound to regulatory proteins |
|
|
What is determin Promoter Proximal Elements? |
Cell needs (E) Conditions (T) |
|
|
Tissue specific transcription factors |
Differential expression of transcription factors determine gene expression according to cells type |
|
|
Unlike bacteria, transcription in eukaryotes does not involve
|
operons
|
|
|
instead operon mechanism in EK |
, the genes encoding enzymes that participate in thesame mechanism are scattered indifferent chromosomes, but they have the same promoter proximal elements and activate at same time in harmony |
|
|
cis-acting regulatory sequences
|
enhancers
|
|
|
e binding sites for gene-specifictranscription factors that regulate RNA polymerase II such as aprotein called the Mediator
|
enhancers |
|
|
The ability of enhancers tofunction even whenseparated by long distancesfrom transcription initiationsites is possible because of
|
DNA looping.
|
|
|
enhancers can regulate transcriptionregardless of orientation or location because of |
DNA looping – as long as there’s no mutations.
|
|
|
work the sameway enhancers do(through DNA looping),except they represstranscription instead ofstimulating it.
|
silencers |
|
|
Mechanism ofTranscription
|
1. Initiation: 2. Elongation: 3. Termination: |
|
|
initiation mechanism |
• The first step of initiation is the binding of a general transcriptionfactor called TFIID to the promoter.
• The binding is followed by recruitment of other proteins to thepromoter and formation of the pre-initiation complex. • Some of the aforementioned recruited proteins are RNApolymerase II and TFIIH. • TFIIH can function as a helicase and unwind the DNA around theinitiation site, exposing the DNA template to RNA polymerase andcreating an open promoter complex. |
|
|
• SNPs in the promoter region can |
affect the binding oftranscription factors required for expression of genes
|
|
|
latent SNPs can lead to |
increase or a decrease of theexpression of the affected gene, which eventually can influencethe risk of developing a disease so it affects the phenotype.
|
|
|
Movement of RNA polymerase and beginning of elongation iscatalyzed by
|
TFIIH
|
|
|
Movement of RNA polymerase and beginning of elongation iscatalyzed, again, by TFIIH--- why
|
TFIIH has protein kinase subunits, which means it can alsofunction as kinase andphosphorylate the tail ofRNA polymerase. This allows the RNApolymerase to thendissociate and moveforward.
|
|
|
Termination is determined by
|
consensus sequence for termination
|
|
|
The consensus sequence for termination |
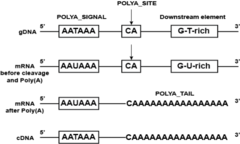
AAUAAA, followed by CA, and 10- 30 nucleotides downstream by a GU-richsequence. |
|
|
why EK gene termed monocistronic |
Eukaryotic transcription units or genes produce mRNAs that encode only one polypeptide at a time
|
|
|
introns |
non-coding specific DNA sequences |
|
|
The protein-coding regions are called
|
|
|
|
When the RNA is synthesized, it contain
|
both introns and exons and is known as pre-mRNA.
|
|
|
Significance of Introns:
|
• They can encode functional RNAs such as nucleolar RNA (that functions in ribosomal processing) as well as microRNAs.
• They contain regulatory sequences of gene expression, such as enhancers. • Variation among individuals can be generated via genetic recombination. • The exon-intron arrangement may facilitate the emergence of new proteins via alternative splicing. |
|
|
how introns are affect Genetic recombination |
crossing over between exons occurs. will result in the production of different germ cells. • For example, the resulting germ cell may have one exon from each parent, and it also may have two exons from the same parent, and so on.
|
|
|
alternative splicing is |
It is tissue specific
|
|
|
alternative splicing results in
|
produce different mRNAs and different proteins isoforms
|
|
|
tropomyosin is an actin binding protein found in muscle cells. Its regulation and activity level is different among different types of muscle cells so there are different isoforms of this protein in each type of muscle cell
|
This happens through alternative splicing.
|
|
|
alternative splicing |
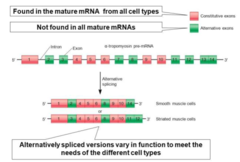
|
|
|
How different cells appear differnet in mRNA? |
They synthesize different sets of RNA and protein molecules |
|
|
Are mRNA patters can differentiate cell types from each other |
Jaaa |
|
|
What we use in isolation of transcreption factors ? |
Affinity chromatography
|
|
|
How we identify DNA sequence to which a Transcription Factor binds ? |
Chromatin Immunoprecipitation |
|
|
Northeern Blotting Use for |
To know the sequence of nucleotides in mRNA so we know the sequence of Amino Acids in that gene (codon) |
|
|
In situ Hybridization methods proprites |
Reveals the distribution of specific RNA molecules in cells in tissue |
|
|
In situ hyberidization incubated by |
Complementary sequence of target sequence of DNA or RNA probe |
|
|
In situ hybredization benefits |
1.Observed of differential gene expression pattrens . 2.Location of RNAs can be determined in cells . |
|
|
DNA library is for |
DNA fragments |
|
|
DNA library types |
cDNA library Genomic library |
|
|
Differnce between Genomic and cDNA libraries ? |
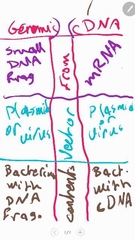
|
|
|
Is we use Reverse transcriptase withe ..... Genomic cDNA LIBRARIES |
Geenomic no cDNA yes |
|
|
How q real-time PCR taken place |
Converting RNA into cDNA followed by PCR in the presence of SYBR green |
|
|
Quantitative of RNA methods |
q-PCR Nothern Blotting |
|
|
Nothern blotting can deduce alot of information .what is it ? |
No. Of RNA in sample Length of it Sequences of RNA in sample |
|
|
What is the criterion in qPCR |
The higher amount of RNA cDNA the sooner it detected |
|
|
DNA Microarrays is used in |
Studying of Transcriptomes |
|
|
The benefet from studying the transcriptome |
Understand gene expression patterns in physiological and pathological states |
|
|
DNA Microarray is done for |
A single sample using radioactive labeled cDNA |
|
|
To compare expression of genes two different samples we use |
Comparative Expression |
|
|
To differentiate the cDNA from each samples we |
Fluorescently labeled it with different colors and added it to array |
|
|
Protein sythesis involves interaction between ? |
tRNA rRNA mRNA |
|
|
What is an operon |
A cluster of genes transcribed from one promoter producing a polycistronic mRNA . |

