![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
19 Cards in this Set
- Front
- Back
|
Paracrine Signaling |
Cell secretes signaling molecules that induce transduction pathways in nearby cells
Signal molecule does not need to travel through blood to reach target cells |
|
|
Autocrine Signaling |
Cell secretes signaling molecules that bind to autocrine receptors on its own plasma membrane |
|
|
Synapse Signaling |
Involves pre-synaptic and post-synaptic cell
Presynaptic cell releases signaling molecule into intercellular space
Signaling molecule binds to receptor on plasma membrane of post-synaptic cell and induces cell signaling pathway |
|
|
Endocrine Signaling |
Cell secretes signaling molecules that travel through the blood stream to reach target cells |
|
|
List the major types of plasma membrane receptor and illustrate how they are activated |
1. Ion Channel Coupled Receptors - signal molecule binds to receptor and opens channel
2. Enzyme Coupled Receptors - signal molecule binds to receptor; enzymatic activity phosphorylates receptor creating binding sites that additional proteins can attach to
3. G Protein Coupled Receptors - signal binds to receptor causing conformational change that activates G protein which induces signal - does not require phosphorylation |
|
|
Predict how mutations affecting the structure of a receptor would affect its functioning |
If the mutation affects the conformation of the receptor's active site, the receptor will no longer recognize its intended signal and will be rendered non-functional
|
|
|
Connect the following terms (time and space): stimulus, ligand, receptor, second messenger, signal transduction, effector response, amplification, cross talk |
1. Ligand (extra cellular space) binds to 2. Receptor (plasma membrane) and acts as 3. Stimulus (plasma membrane) which induces 4. Signal Transduction (cytosol) that activates a 5. Second Messenger (cytosol) which creates 6. Amplification (cytosol) of the signal. 7. Cross Talk (cytosol) may lead to a change in 8. Effector Response |
|
|
List the types of second messenger systems and explain how they work [cAMP] |
*
Adenylyl cyclase (aka adenylate cyclase) Cleaves ATP in AMP and pyrophospate; cAMP phosphodiesterase creates bond between remaining phosphate and 3’ OH Ribose into cAMP * Inactive protein kinase A consists of four subunits (2 regulatory or R subunits and 2 catalytic or C subunits) * cAMP binds to R subunits and causes them to dissociate from C units which are now active * PKA can cause short term changes by phosphorylating T and S residues on proteins * PKA can cause long term changes by activating CREB (cAMP response binding protein) * CREB interacts with cAMP response element which effects transcription * Signal is terminated by cyclic AMP phosphodiesterase by breaking the ring and converting the molecule to AMP |
|
|
List the types of second messenger systems and explain how they work [lipids] |
* PI is on inner leaflet of the plasma membrane inside because we need it there for signaling Phopholipase C cleaves phosphotidlyinositol 4,5-bisphosphate (PIP2) into diacylglycerol (DAG) and inositol 1,4,5-trisphosphate IP3 * IP3 goes on to transduce the a signal. Often leads to stimulation of the smooth ER to release Ca++ ions * PTEN terminates the signal by converting IP3 back into PIP2 and phosphate - works as a tumor suppressor because its blocking mechanisms that are needed to proliferate |
|
|
List the types of second messenger systems and explain how they work [Ca²⁺] |
Ca²⁺ ions released from smooth ER bind to calmodulin and then the complex binds to the inactive form of Ca²⁺/calmodulin-dependent protein kinase thus activating it
|
|
|
Define the role of G proteins in cell signaling |
G proteins act as molecular switches that induce transduction of cell signals.
They are activated by a conformational change that occurs when the receptor they are complexed with binds a liganda |
|
|
Tyrosine Receptor Kinase [Ras Dependent Signaling] |
ras-dependent pathway - growth factor comes and helps the receptor dimerize (exception = insulin) - you have cross phosphorylation and autophosphorylation - this creates docking sites - the docking sites attract the adaptor protein (in this case Grb2) - one of the mechanisms is for the adaptor proteins to attract SOS (gef), the exchange factor that exchanges GDP for GTP Ras-GDP binds to SOS (gef). - this kicks out the GDP and then GTP comes in Ras-GTP is active - once the Ras-GTP is active, it is going to activate other proteins - this can then cause the activation of the MAP kinase pathway |
|
|
Tyrosine Receptor Kinase [MapK Pathway] |
MAPK pathway - activated Ras-GTP will phosphorylate and activate a Raf (which is a MAP kinase-kinase kinase) - activated Raf will then phosphorylate and activate MEK (which is a MAP kinase-kinase) - activated MEK will then phosphorylate and activate ERK (which is a MAP kinase) - through the cascade of phosphorylation you will eventually have a map kinase that can enter the nucleus and increase transcription, thus activating proteins these proteins will then have a biological action like cell proliferation etc. |
|
|
Tyrosine Receptor Kinase [PI3K/AKT Pathway] |
Ras Independent Signaling - signal binds and helps the receptor dimerize - you have cross phosphorylation and autophosphorylation - this creates docking sites - the docking sites attract the adaptor protein (IP3 kinase in this case) - IP3 kinase phosphorylate PI 4,5-bisP creating PI 3,4,5-trisP - PI 3,4,5-trisP serves as docking site that activates Protein Kinase 3 or AKT |
|
|
Serine/Threonine Receptor Kinase [TGF-β] |
Example: TGF-beta binds to the receptor - in this case, TGF-beta binds to a type II receptor - type II receptor recruits a type I receptor - type II then phosphorylates type I, which creates docking sites for 2 R-Smad (regulatory SMAD) - Both R-Smads are then phosphorylated, allowing them to bind to 1 Co-Smad - this exposes the localization signal on co-smad which allows R-smad and co-smad to migrate to the nucleus and regulate gene expression
|
|
|
JAK/STAT Receptor |
- similar in that they form a homodimer - the binding of the ligand will bring these two together - receptor doesn’t have enzymatic activity but it is bound to an enzyme (JAK) that phosphorylates it - this phosphorylation creates a docking site to which the STAT protein can bind, - STAT is then phosphorylated by JAK and detaches from the receptor - It then homodimerizes with another phophorylated STAT and translocates to the nucleus where it promotes transcription |
|
|
G Protein Coupled Receptor |
Do not have any enzymatic activity The G protein is in it’s “off state” with GDP bound to its Gα subunit When the ligand comes, there is a conformational change that activates the receptor Once the conformational change occurs, GDP can then be exchanged for a GTP on the Gα subunit This then triggers the dissociation of the Gα subunit bound to GTP from the G,β,δ dimer and the G receptor Both Gα-GTP and Gβδ can then activate different signaling cascades (or second messenger pathways) and effector proteins, while the receptor is able to activate the next G protein Gα subunit will eventually hydrolyze the attached GTP to GDP by its inherent enzymatic activity, allowing it to re-associate with G,β,δ and starting a new cycle. A group of proteins called Regulator of G protein signalling (RGSs), act as GTPase-activating proteins (GAPs), specific for Gα subunits. These proteins act to accelerate hydrolysis of GTP to GDP and terminate the transduced signal
|
|
|
Recognize the importance and general mechanisms of signal termination |
Signal needs to be terminated so the effects they cause do not continue for ever
In most cases an enzyme deactivates one of the proteins that the receptor activated
ex; GRS dephosphorylates α-GTP to α-GDP which then recomplexes with the β and δ subunits ending the signal transduction pathway |
|
|
Contrast intracellular and extracellular receptors and illustrate with examples |
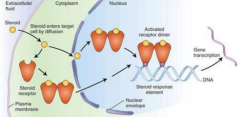
Hydrophilic signals bind to extracellular receptors
Hydrophobic ones traverse membrane and bind to intracellular ones
Intracellular receptors can be in the cytosol or in the nucleus |

