![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
182 Cards in this Set
- Front
- Back
|
Alanine
|
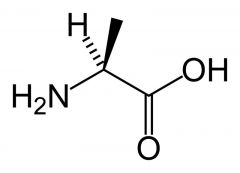
What is the name of this amino acid?
|
|
|
Arginine
|
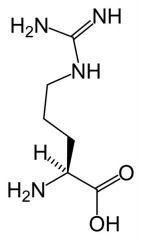
What is the name of this amino acid?
|
|
|
Asparagine
|

What is the name of this amino acid?
|
|
|
Aspartate (Aspartic acid)
|

What is the name of this amino acid?
|
|
|
Cysteine
|
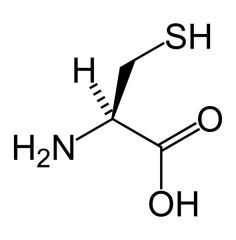
What is the name of this amino acid?
|
|
|
Glutamate (Glutamic acid)
|
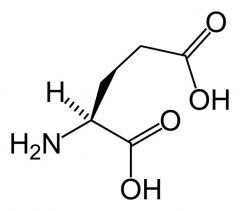
What is the name of this amino acid?
|
|
|
Glutamine
|
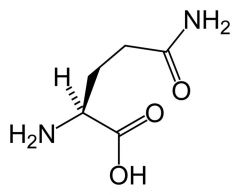
What is the name of this amino acid?
|
|
|
Glycine
|
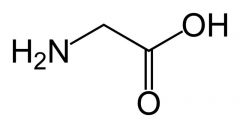
What is the name of this amino acid?
|
|
|
Histidine
|
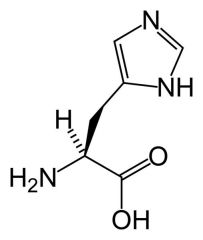
What is the name of this amino acid?
|
|
|
Isoleucine
|
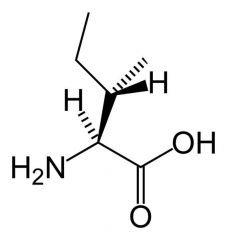
What is the name of this amino acid?
|
|
|
Leucine
|
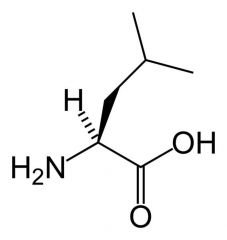
What is the name of this amino acid?
|
|
|
Lysine
|
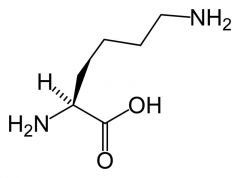
What is the name of this amino acid?
|
|
|
Methionine
|
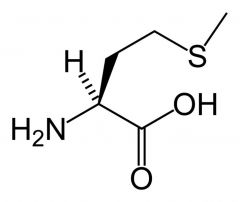
What is the name of this amino acid?
|
|
|
Phenylalanine
|
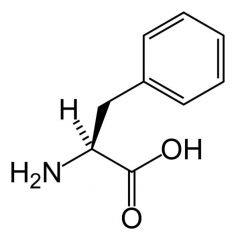
What is the name of this amino acid?
|
|
|
Proline
|
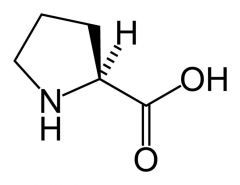
What is the name of this amino acid?
|
|
|
Serine
|
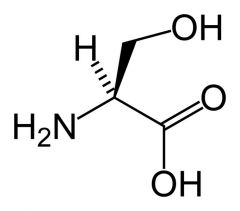
What is the name of this amino acid?
|
|
|
Threonine
|

What is the name of this amino acid?
|
|
|
Tryptophan
|
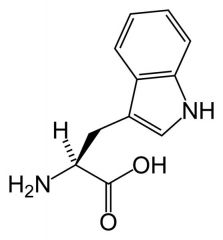
What is the name of this amino acid?
|
|
|
Tyrosine
|

What is the name of this amino acid?
|
|
|
Valine
|
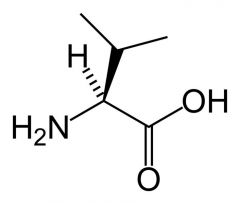
What is the name of this amino acid?
|
|
|
Glycine
|
*Nonpolar/Aliphatic*
Gly G |
|
|
Alanine
|
*Nonpolar/Aliphatic*
Ala A |
|
|
Proline
|
*Nonpolar/Aliphatic*
Pro P |
|
|
Valine
|
*Nonpolar/Aliphatic*
Branched Chain Val V |
|
|
Leucine
|
*Nonpolar/Aliphatic*
Branched Chain Leu L |
|
|
Isoleucine
|
*Nonpolar/Aliphatic*
Branched Chain Ile I |
|
|
Phenylalanine
|
*Aromatic*
Nonpolar Phe F |
|
|
Tyrosine
|
*Aromatic*
More polar Tyr Y |
|
|
Tryptophan
|
*Aromatic*
More polar Trp W |
|
|
Asparagine
|
*Polar, Uncharged*
Asn N |
|
|
Glutamine
|
*Polar, Uncharged*
Gln Q |
|
|
Serine
|
*Polar, Uncharged*
Ser S |
|
|
Threonine
|
*Polar, Uncharged*
Thr T |
|
|
Methionine
|
*Sulfur Containing*
Met M |
|
|
Cysteine
|
*Sulfur Containing*
Cys C |
|
|
Aspartate
|
*Charged*
Negative (Acidic) Asp D |
|
|
Glutamate
|
*Charged*
Negative (Acidic) Glu E |
|
|
Arginine
|
*Charged*
Positive (Basic) Arg R |
|
|
Lysine
|
*Charged*
Positive (Basic) Lys K |
|
|
Histidine
|
*Charged*
Positive (Basic) His H |
|
|
phosphorylation targets?
|
ser
thr |
|
|
Met is nearly always associated with ____________ of proteins
|
hydrophobic cores
|
|
|
What amino acids account for strong absorption at 280nm?
|
aromatics:
phe tyr trp |
|
|
henderson hasselbalch equation?
|
pH = pKa + log (base/acid)
|
|
|
Approximate pKa for N-terminal? C-terminal group?
|
N-terminal = ~7.6-10.6
COOH = ~3.0-5.5 |
|
|
Approximate pKa for R group of:
arg |
12.5
|
|
|
Approximate pKa for R group of:
cys |
8.5
|
|
|
Approximate pKa for R group of:
His |
6
|
|
|
Approximate pKa for R group of:
tyr |
10
|
|
|
Approximate pKa for R group of:
asp |
3.8
|
|
|
Approximate pKa for R group of:
glu |
4.2
|
|
|
Approximate pKa for R group of:
lys |
10
|
|
|
alpha helix
phi =? psi=? |
phi = -57.8
psi = -47 |
|
|
average alpha helix length (# of res)?
|
10 residues
|
|
|
alpha helix
residues per turn (crystallographic repeat)? |
c = 18 residues (5 x 3.6)
5 turns per repeat |
|
|
alpha helix residues per turn?
pitch? rise? |
3.6 residues per turn
pitch = 0.54nm rise = 0.15nm/residue |
|
|
The hydrogen bonding takes place between the carbonyl group of residue I and the hydrogen from the amide group ____ residues ahead
|
The hydrogen bonding takes place between the carbonyl group of residue I and the hydrogen from the amide group four residues ahead (i+4)
|
|
|
The most stretch helix will have the highest ____ and the lowest ________
|
The most stretch helix will have the highest rise and the lowest number of residues per turn
|
|
|
B-sheets are composed of __ or more different
stretches of at least ___to___ amino acids. |
B-sheets are composed of 2 or more different
stretches of at least 5-10 amino acids. |
|
|
residues per "turn" in a B-sheet?
phi =? psi = ? |
n = 2 residues per turn
phi = -120 psi = +120 |
|
|
distance between residues in B sheet for
anti-parallel? parallel? |
antiparallel = 3.2A
parallel = 3.4A |
|
|
average length of a B-sheet?
|
6 residues
|
|
|
Turns are located primarily _______ and contain ________ residues
|
Turns are located primarily on the protein surface and contain polar and charged residues
|
|
|
Loops are located primarily _______ and contain ________ residues
|
Loops are located primarily on the protein surface and contain polar and charged residues
|
|
|
primary role of alpha-1-antitrypsin? why?
|
inhibit neutrophil elastase so it doesn't break down elastic fibers of the lungs
|
|
|
S mutation of alpha-1-antitrypsin
|
Glu264 to Val
|
|
|
Z mutation of alpha-1-antitrypsin
|
Glu 342 to Lys
|
|
|
hallmark of Z alpha-1-antitrypsin liver disease?
|
acid-Schiff-positive inclusion bodies
|
|
|
inactivation of neutrophil elstase occurs when
|
inactivation of neutrophil elstase occurs when it moves from upper to lower pole of A-1-antitryp
|
|
|
what major conformational change occurs in prion disease
|
alpha helixes are converted to beta sheets
|
|
|
fractional saturation of mgb equation?
|
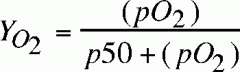
|
|
|
Helix of the histidine that ligates heme( the 5th coordination site)?
residue # in mgb, b-chain, a-chain? |
Helix F8
mgb = residue 93 hgb b-chain = residue 92 hgb a-chain = 87 |
|
|
6th coordination site in the deoxy state? helix and residue numbers?
|
helix E7
mgb = residue 64 hgb b-chain = residue 63 hgb a-chain = residue 58 |
|
|
fractional saturation of HGB
|
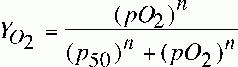
|
|
|
Hill equation of HGB
|
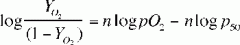
|
|
|
The _____ subunits of hgb may or may not be identical while the ____ subunits are identical
|
alpha = maybe identical
beta = identical |
|
|
R state = ?
T state = ? |
R state = relaxed = oxy
T state = taut = deoxy |
|
|
What residue facilitates the rotation of the alpha/beta dimer when o2 binds?
|
His F8
|
|
|
Deoxy/T-state stabalizing interactions?
|
-C-terminius of Beta HIS lies on top of helix C in alpha and interacts with Asp 94 of the same β2 by charge-charge interaction and with Lys 40 of the α1 through charge-charge interactions
-N-terminal (NH3 grp) residue of α1 interacts with the guanidinium group of carboxyl terminal residue (Arg) of α2 via charge-charge + Cl- -guanidinium group of the C- terminal residue Arg141 of the α1 interacts with Asp126 of α2 -Tyr 140 of α1 hydrogen bonds to the carbonyl group of Val 93 in the same subunit. Similarly, Tyr145 in the β subunit forms a hydrogen bond with the carbonyl group of Val98. |
|
|
Bohr effect in terms of specific residue
|
excess protons protonate C terminus B2 146 His and stabalize the deoxy/T state
|
|
|
where does carbamation happen? effect?
|
carbamation happens N-terminus and stabalizes the salt bridge formation between alpha and beta chains
|
|
|
what residue is replaced by ser in fetal gamma subunit? effect?
|
his 143 is replaced by ser in fetal gamma subunit = positive charge replaced by negative = repels BPG = fHgb has less affinity for BPG = fHgb has higher O2 affinity
|
|
|
amino acid residues involved in binding 2,3BPG are
|
Lys82, His143, His2, and the N-terminal amino group of the β- chains?
|
|
|
substitution in sickle cell
|
β6 Glu -> Val
|
|
|
Sickle cell hgb on electrophoresis?
|
runs slower; close to the anode
|
|
|
Codes for Glu and Val
|
Glu = GAG
Val = GTG |
|
|
β6 Glu -> Ala
|
• Changes in helix A of β6 from Glu to Ala produces insignificant sickling. Ala, due to its small size,
probably does not fit in the hydrophobic EF pocket of the β chain very well. |
|
|
β121 Glu -> Lys
|
• Enhanced sickling
• The reason for this enhanced sickling is that switching from Glu (negative charge) to Lys (positive charge) causes charge- charge interactions with β6 Glu. |
|
|
His, F8, αchain (α87) -> Tyr
|
• Replacement of His with Tyr results in ligating the ferrous atom in the heme and the Fe+2 becomes susceptible to oxidation to the ferric state (Fe+3)and no longer binds oxygen, resulting in reduced oxygen binding affinity.
|
|
|
Arg 141 (α chain) -> His
|
This variant is characterized by increased oxygen affinity (smaller p50) favoring the R state.
This is attributed to the elimination of the interactions between Arg and Asp126 of α chain in deoxyHb. |
|
|
• β Gly74 -> Asp.
|
introducing a negatively charged residue in this region reduces the binding of 2,3-bisphosphoglycerate to Hb resulting in higher affinity Hb.
|
|
|
• β His146 -> Asp
|
Since this residue is critical for the Bohr effect and it plays a role in deoxy to Oxy Hb switching, the changes to Asp results in formation of Hb with high affinity for oxygen
|
|
|
β His (F8) -> Gln
|
This variant does not hold on to its heme because Gln does not coordinate well with the ferrous atom in the heme, opening the hydrophobic crevice for polar solvent.
|
|
|
β 42 Phe -> Ser
|
• This variant is unstable and loses its heme.
• replacement of the more hydrophobic amino acid residue Phe with Ser, which is hydrophilic and polar, opens the pocket for water and results in loss of heme. |
|
|
Pro (α chain) -> Arg
|
• The loss of Pro results in a change of the geometry of the subunit and therefore alters subunit interactions due to continuation of the helix. This results in Hb dissociation into subunits.
|
|
|
Leu136 (α chain) -> Pro
|
Since introduction of proline into the helix interrupts the helix, it results in dissociation of the tetramer and results in high oxygen binding affinity.
|
|
|
alpha genes are on chromosome?
beta genes are on chromosome? |
alpha = 16
beta = 11 |
|
|
How many residues in alpha chain of HGB?
How many residues in beta chain of HGB? |
alpha = 141
beta = 146 |
|
|
the plane of ______ and ______ is rotated to avoid collision with the heme
|
valFG5 and leuFG3
|
|
|
relative rotation of alpha-beta units in hgb?
|
15degrees
|
|
|
His F8 function in hgb?
|
proximal his that binds Fe(II)
|
|
|
His E7 function in hgb?
|
Distal histidine, protects the heme and forces O2 to bind at an angle
|
|
|
Phe CD1 function in hgb?
|
Heme contact
|
|
|
Leu F4 function in hgb?
|
Heme contact
|
|
|
Gly B6 function in hgb?
|
Allows approach of B and E helices
|
|
|
Pro C2 function in hgb?
|
Terminates the helix
|
|
|
Tyr HC2 function in hgb?
|
hydrogen bond between H and F helices
|
|
|
On assuming the R state the iron moves down ____ angstroms into the same plane as the porphyrin.
|
0.6A
|
|
|
Enzyme classification number:
First number? Fourth number? |
first = n1 = type of reaction
last = n4 = substrate |
|
|
Enzyme classifications:
n1 = 1 |
Oxidoreductase
Oxidation-reduction: Transfer of electrons from one molecule (donor, reductant) that is oxidized to another (acceptor, oxidant) that is reduced |
|
|
Enzyme classifications:
n1 = 2 |
Transferase
Transfer of functional group (C-, N-, P-containing) from one molecule to another |
|
|
Enzyme classifications:
n1 = 3 |
Hydrolase
Cleavage of bonds by hydrolysis |
|
|
Enzyme classifications:
n1 = 4 |
Lyase
Addition or removal of groups to form double bonds |
|
|
Enzyme classifications:
n1 = 5 |
Isomerase
Isomerization: Intramolecular group transfer |
|
|
Enzyme classifications:
n1 = 6 |
Ligase
Joining two molecules by covalent bonds with breakdown of ATP |
|
|
Cosubstrate is _______ by a rxn
prosthetic group is _________ by a rxn |
Cosubstrate is loosely bound and changed by a rxn
prosthetic group is tightly or covalently bound and unchanged by a rxn |
|
|
standard free energy change equation
|
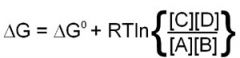
|
|
|
Chymotrypsin cleaves peptide bonds selectively on the ______ side of _________ amino acids (methionine)
|
Chymotrypsin cleaves peptide bonds selectively on the carboxyl side of aromatic (tryptophan, tyrosine, phenylalanine ) and large hydrophobic amino acids (methionine)
|
|
|
Chymotrypsin Catalytic triad?
|
Ser 195, His 57, Asp 102
|
|
|
michaelis-menten equation
|
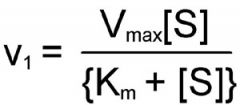
|
|
|
active sites filled equation?
|
[ES]/[E]t = v/vmax = [S]/Km+[S]
|
|
|
When [S] is >>>> Km?
[S] <<< Km |
S > Km = zero order; usually [S] = 20Km
S <<<Km = zero order V proportional to [S] |
|
|
When [S] = Km?
|
When [S] = Km:
v = 0.5vmax |
|
|
Michaelis-Menten 5 assumptions?
|
1) Formation of ES complex between enzyme and substrate
2) No back reaction from product buildup: k4 = 0 at t ~ 0 3) Initial velocities used for analysis (t ~ 0) 4) Steady-state assumption for [ES] = [ES] doesnt change 5) Negligible depletion of substrate: [S] >> [E] |
|
|
Small Km = ?
|
Small Km = high affinity
Large Km = low affinity |
|
|
catalytic efficiency?
|
kcat/Km
You always want the highest kcat |
|
|
Lineweaver Burk plot equation
|
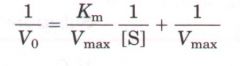
|
|
|
Competitive Inhibition:
Binds? Ki = ? Km, app = ? Vm, app =? Effect on Km? Vmax? catalytic efficiency? |
Binds only to active site
Ki = [E][I]/[EI] Km, app = 1 + ([I]/Ki) Km increases, Vmax doesn't change, cat-eff decreases |
|
|
Uncompeitive Inhibition:
Binds? Ki = ? Km, app = ? Vm, app =? Effect on Km? Vmax? catalytic efficiency? |
Binds only ES complex
Ki = [ES][I]/[ESI] Km, app = Km/{1 + ([I]/Ki)} Vm,app = Vm/{1 + ([I]/Ki)} Km decreases, Vmax decreases, cat-eff no change |
|
|
Noncompeitive Inhibition:
Binds? Ki = ? Km, app = ? Vm, app =? Effect on Km? Vmax? catalytic efficiency? |
Binds to E or ES at a place different than active site
Ki = [E][I]/[EI] = [ES][I]/[ESI] Km, app = Km Vmax, app = Vm/{1 + ([I]/Ki)} Km doesnt change, Vm decreases, Cat-eff decreases |
|
|
succinyl CoA + glycine --> ? enzyme?
|
succinyl CoA + glycine --> delta aminolevulinic acid (ALA)
ALA synthase |
|
|
ALA x 2 --> ? enzyme?
|
ALA x 2 --> porphobilinogen (PBG)
ALA dehydrase |
|
|
PBG x 4 --> ? enzyme?
|
PBG x 4 --> hydroxymethyl bilane (bilane)
PBG deaminase |
|
|
bilane --> ? enzyme?
|
bilane --> uroporphyrinogen III (uro’gen III)
urogen III cosynthase |
|
|
uro’gen III --> ? enzyme?
|
uro’gen III --> coproporphyrinogen III (copro’gen III)
uro’gen III decarboxylase |
|
|
copro’gen III --> ? enzyme?
|
copro’gen III --> protoporphyrinogen IX (proto’gen IX)
copro’gen III decarboxylase |
|
|
proto’gen IX --> ? enzyme?
|
proto’gen IX --> protoporphyrin IX (proto IX)
proto’gen dehydrogenase |
|
|
proto IX --> ? enzyme?
|
proto IX --> heme
ferrocheletase |
|
|
Starting reactants in order for Heme synthesis?
|
1. succinyl CoA + glycine
2. ALA x 2 3. PBG x 4 4. bilane 5. uro’gen III 6. copro’gen III 7. proto’gen IX 8. proto IX |
|
|
Acute Intermittent Porphyria (AIP)? Enzyme defect? Step? Photosensititve?
|
Step 3 = PBG deaminase
NOT photosensitive |
|
|
Hereditary Coproporphyria (HC) Enzyme defect? Step? Photosensititve?
|
Step 6 = Coprogen III Decarboxylase
PHOTOsensitive |
|
|
Porphyria Cutanea Tarda (PCT) Enzyme defect? Step? Photosensititve?
|
Step 5 = Urogen III Decarboxylase
PHOTOsensitive |
|
|
Variegate Porphyria (VP) Enzyme defect? Step? Photosensititve?
|
Step 7 = Protogen IX Dehydrogenase
PHOTOsensitive |
|
|
Urine/Stool findings for the Acute Porphyrias?
Treatment? |
Rx = IV Heme
Urine: ALA PBG Uro Copro Stool: Copro Proto |
|
|
Factors associated with PCT?
|
• Chronic hepatitis C infection
• Alcohol use • Iron overload (Increased hepatic iron inhibits UROD activity) • HIV infection |
|
|
Diagnostic signs for PCT
|
• Characteristic rash
• Increased uroporphyrin int he urine • Emission peak on plasma flourescence • Check for genetic hemochromatosis |
|
|
treatment for PCT?
|
Based on cause:
-Chloroquinone -Blood draw -No sun or alcohol -Treat HepC |
|
|
1/3 of all amino acid residues in collagen are?
|
glycine
|
|
|
1/4 of all amino acid residues in collagen are?
|
proline
|
|
|
1/2 of all proline residues are
|
hydroxylated at carbon 3 or 4 to form hydroxyproline
|
|
|
The repeating polypeptide of collagen is Gly-X-Y where X is usually? and Y is usually?
|
X is usually Pro
Y is usually hydroxyproline |
|
|
Polarity of Gly-Pro-Y and Gly-X-Y
|
Gly-Pro-Y is usually non-polar
Gly-X-Y is most often polar |
|
|
Microheterogeneity
|
different parts of the peptided are hydroxylated
|
|
|
MW of collagen is 300,000 Da, how many residues? residues/chain?
|
3,000 residues
1,000 residues/chain |
|
|
Proline helix?
|
Type II trans helix = left handed polyproline helix with pyrrolidine rings
|
|
|
poly-proline helix:
rise? |
2.8A
|
|
|
Tight binding of alpha chains is facilitated by
|
Gly at every 3rd position
|
|
|
Function of N-terminal leader sequence in transcripted alpha chain>
|
signals translation by rER ribosomes
|
|
|
Enzyme that post-translationally modifies proline residues? cofactor? primarily at what carbon? where in cell?
|
prolyl hydroxylase hydroxilates mainly at C4
cofactor = ascorbate in the ER |
|
|
Enzyme that hydroxylates lys? what carbon? where do hydroxylysines usually appear in the collagen polymer?
|
Lysyl hydroxylase
5th carbon Hydroxylysines frequently occur at the Y-position in the sequence (Gly-X- Y) |
|
|
Unique reactant needed for prolyl and lysyl hydroxylase?
byproducts? |
reactant O2 (and ascorbate)
byproducts = succinate and CO2 |
|
|
Once in the __________, some _________ residues are glycosylated with galactose monosaccharides or glucosylgalactose disaccharides.
|
Once in the Golgi apparatus, some hydroxylysine residues are glycosylated with galactose monosaccharides or glucosylgalactose disaccharides.
|
|
|
Does procollagen have disulfide bonds?
|
Yes in the C-terminal extensions
|
|
|
Does tropocollagen have disulfide bonds?
|
usually no
|
|
|
Tropocollagens align with ___ stagger in a non-symmetrical way with ____nm gaps between tropocollagens. Adjacent rows are displaced by ___nm, and the structure repeats every ___ rows.
|
Tropocollagens align with 1⁄4 stagger in a non-symmetrical way with 40nm gaps between tropocollagens. Adjacent rows are displaced by 68nm, and the structure repeats every 5 rows.
|
|
|
Fibers are stabilized by the covalent cross-linking of the constituent collagen molecules. The cross-links result from the ________ in lysine (or hydroxylysine) into an _______ by the enzyme ________
|
Fibers are stabilized by the covalent cross-linking of the constituent collagen molecules. The cross-links result from the oxidation of the amino group in lysine (or hydroxylysine) into an aldehyde by the enzyme lysyl oxidase (LOX).
|
|
|
hydroxypyridinium ring (pyridinoline) forms between ?
|
two hydroxylysyl and one lysyl residues.
|
|
|
BAPN?
|
suicide inhibitor of LOX, and the resulting reduced cross-linkage of collagen fibers caused by BAPN is reflected by severe abnormalities in the physiology of bones, joints and blood vessels.
|
|
|
MMPs?
|
matrix metalloproteinase’s (MMPs) enzymes cleave the collagen triple helix about three quarters from the N-terminus, thus decreasing the Tm from slightly above 37°C to about 30°C.
|
|
|
elastin is a ____ molecule and is ___ glycosylated. About one third of the amino acid residues are ____, and about one half are ___
|
elastin is a single molecule and is not glycosylated. About one third of the amino acid residues are Gly, and about one half are Pro
|
|
|
do gly and pro elastin form a regular secondary structure
|
no
|
|
|
Each molecule is about 70kDa and contains_______ regions that participate in _______ interactions.
|
70,000Da (~100 residues)
valine-rich non-polar regions that participate in hydrophobic interactions. |
|
|
Function of lysyl oxidase on elastin?
|
cross-links to form highly insoluble elastin fibres
|
|
|
cross-linking requires lysyl oxidase to form ________ residues that react with the _______, unaltered lysine residue to form a _______
|
cross-linking requires lysyl oxidase to form three allylysine residues that react with the amino group of a fourth, unaltered lysine residue to form a desmosine
|
|
|
Prolyl hydroxylase example of disease?
|
scurvy
|
|
|
Lysyl hydroxylase example of disease?
|
Ehlers-Danlos type VI
|
|
|
Lysyl oxidase example of disease
|
Osteolathyrism Copper deficiency Vitamin B6 deficiency Menkes kinky-hair syndrome D-Penicillamine Ehlers-Danlos type V
|
|
|
Requirements for the function of prolyl hydroxilase?
|
ascorbate
alpha ketoglutarate Molecular O2 Ferrous |
|
|
Requirements for function of lysyl hydroxylase?
|
ascorbate
alpha ketoglutarate Molecular O2 Ferrous |
|
|
Requirements for function of Lysyl oxidase
|
Copper
Pyridoxal phosphate Molecular oxygen |
|
|
Proteoglycans are extracellular proteins with covalently bound glycosaminoglycans (GAGs) O-linked to _____
charge? |
serine residues.
very negative |
|
|
Is hyaluronic acid linked to proteins?
|
no
it is the exception of proteoglycans |

