![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
115 Cards in this Set
- Front
- Back
|
What are pAMP and pKAN? |
Plasmids, each carrying the ampicillin and kanamycin-resistance and only those genes, respectively. |
|
|
Describe the steps necessary for restriction digest of pAMP and pKAN. |
1. To separate pAMP and pKAN samples, add 2x restriction buffer. 2. Expose separate samples of pKAN and pAMP to the enzymes BamHI and HindIII. 3. Incubate at 37C (allows for digestion to occur). 4. Perform gel electrophoresis of digested samples, control pAMP, control pKAN, and Lambda DNA Maker digested BstEII. 5. |
|
|
Into what fragments are pAMP and pKAN digested? What is the simplest recombinant plasmid like? |
pAMP: 3755bp fragment with amp and ORI; a 784bp fragment
pKAN: 1875bp fragment with kan; a 2332bp fragment with ORI
The simplest recombinant has the amp/ORI fragment and kan fragment.
|
|
|
In ligation of pAMP and pKAN, describe the steps. |
1. Heat the digested samples to destroy restriction enzymes. 2. Mix both, along with DNA ligase, water and ATP/2x ligation buffer (powers ligation). 3. Incubate at 25C to allow for ligation to occur. |
|
|
How do hydrogen bonds and phosphodiester bonds form in the ligation process? |
- the sticky ends are single stranded and complement each other, leading to H-bond formation - ligase catalyzes phosphodiester bond formation (covalently link the bases) |
|
|
In electrophoresis of digested pAMP and pKAN, what do fragments that do not correspond to the expected bands mean? |
- incomplete digestion - fix by adding more restriction enzymes and incubation |
|
|
How do you ensure ligation was performed correctly? |
Perform electrophoresis. None of the digested BamHI/HindIII-digested fragments should appear but only (ideally) one large fragment. |
|
|
Many combinations of fragment may come together. Which are these? Which ones will be maintained and expressed? |
- the recombinant plasmid with both resistance genes, regenerated pAMP, regenerated pKAN - only those with ORI |
|
|
In gel electrophoresis, what is used for staining DNA? |
- ethidium bromide; which intercalates into the bases and is seen by photograph - bromophenol blue moves the fastest and is used to track the progress of electrophoresis |
|
|
Describe the process of transformation. |
1. Take ligated plasmid DNA and incubate with E.coli cell suspension at 0C CaCl2 for 20 minutes. 2. Heat shock at 42C. 3. Allow recovery at 37C for 40-60 minutes. |
|
|
Justify the steps of heat shock. |
- Placing the cells in CaCl2 solution at 0C decreases membrane fluidity and the Ca2+ neutralizes the negative charge of DNA. - Heat shock at 42C makes the cell uptake the plasmid. - 37C recovery allows for plasmid DNA to be expressed. |
|
|
For which antibiotic selection is this recovery period most crucial? |
- for kanamycin selection |
|
|
How does ampicillin and ampicillin resistance work? Which enzyme does the ampR gene code for? |
- ampicillin has a B-lactam ring that binds TPase, halting peptidoglycan synthesis - ampR codes for B-lactamase, which gets secreted out of the cell - this enzyme attacks the B-lactam ring in ampicillin |
|
|
How does kanamycin and kanamycin resistance work? |
Kanamycin irreversibly binds 30s subunit of ribosome, halting translation - the kanR product protein is NOT secreted but modifies kanamycin, preventing binding to 30s subunit. |
|
|
What is the overall objective of transformation in terms of this specific experiment? |
- by transforming E.coli cells, we can later select for cells with both resistance genes |
|
|
Who discovered the method for transformation used in this experiment? |
Mandel and Higa |
|
|
At what growth phase are the E.coli cells used in transformation? |
Mid-log phase |
|
|
Stop and review your transformation efficiency. |
OK. |
|
|
Where do you plate your transformed pLIG, pAMP, pKAN cells? |
- LB/amp, LB/kan, LB/amp+kan plates - incubate at 37C for 12-24 hours then at 4C to arrest growth of contaminating organisms |
|
|
Describe sets for preparation of competent cells for transformation. |
1. Obtain mid-log cells. 2. Centrifuge and remove supernatants. 3. Add CaCl2 and finger vortex. Ensure mixture is homogeneous and no clumps visible. 4. Add more CaCl2 and vortex. 5. Return to ice for incubation for 20 minutes. 6. Centrifuge again (CaCl2 alters adhering properties of plasma membrane; cell pellet is dispersed after centrifugation). 7. Remove CaCl2. 8. Re-add new ice-cold CaCl2 and finger vortex. |
|
|
What are expected results for the transformation? Are satellite colonies observed after over-incubation? |
- least colonies on LB/amp+kan (ten-fold less than LB/amp or kan - more colonies in LB/amp than LB/kan
- B-lactamase (ampR product) is secreted, creating a zone of clearance where surrounding area is clear of ampicillin - eventually, even those without ampR gene (including those with kanR only) can grow in this area - kanR is not secreted so no zone of clearance; only ones that grow; also kanR product kills non-resistant cells right away |
|
|
How do you calculate transformation efficiency? |
TE = colony forming units/mass of plasmid spread (ug) |
|
|
Why perform replica plating? |
- We know for sure that those that grew at LB/amp+kan have double resistance but don't know if those that grew in LB/amp and LB/kan are also doubly resistant. - we can learn the success of ligation; usually 30-70% are dual resistant for both LB/amp and kan - methodology: plate LB/amp cells into new LB/amp and LB/kan in same order; do same with LB/kan cells; incubate |
|
|
Design a test to see if ampicillin and kanamycin-resistance gene products are secreted or not. |
1. Grow ampR on LB/amp and kanR on LB/kan. 2. Filter each to remove E.coli cells 3. On a fresh LB/amp plate, divide into two. In top half, add amp filtrate. 4. On fresh LB/kan plate, divide into two. In top half, add kan filtrate. 5. Streak non-transformed E.coli cells on both plates.
On LB/amp plate, E. coli should grow since filtrate had B-lactamase, so no antibiotic. On LB/kan plate, E.coli should not grow since the kanR product was not secreted and was filtered out with E.coli On the untreated sides, E. coli should be observed (control) |
|
|
What was the goal of Lab 4: Purification/Identification of Recombinant DNA? |
To isolate and determine the genotype responsible for dual resistance. |
|
|
Describe mini-prep and justify each step. |
1. Centrifuge culture tubes of transformed E.coli. Remove supernatant (do not disturb pellet). 2. Add GTE buffer (Glucose-TrisHCl-EDTA). EDTA chelates cations used by any present restriction enzymes. 3. Add SDS/NaOH. Mix and place on ice until suspension is clear at about 5 minutes. 4. Add KOAc (Potassium acetate). 5. Centrifuge and remove PRECIPITATE, not supernatant. Recall: we lysed the cell so the DNA is suspended. 6. To the supernatant, add isopropanol and mix. Isopropanol precipitates nucleic acids rapidly and eventually, proteins. So do this step quickly so we only get nucleic acids. 7. Centrifuge and remove supernatant. 8. Add EtOH. 9. Centrifuge and pour off supernatant. The pellet may not be visible but this is a sign of no contamination. Nucleic acids not soluble in EtOH so it will be in pellet. 10. Evaporate remaining EtOH. 11. Resuspend at TE buffer. |
|
|
Purpose of SDS/NaOH. |
Add SDS/NaOH to lyse the cell membrane. It also raises the pH and denatures DNA to sDNA, large RNA is degraded. Circular plasmid DNA remains supercoiled due to intertwining, so they don't get tangled with others larger denatured molecules. Mix and place on ice until suspension is clear at about 5 minutes. |
|
|
Purpose of KoAc. |
It neutralizes pH so DNA is renatured and SDS is precipitated with lipids and proteins. Chromosomal DNA is precipitated because it has protein attachments. |
|
|
Describe steps of restriction analysis of recombinant DNA |
Expose purified plasmids to BamHI/HindIII, water, buffer with RNAse. Perform agarose gel electrophoresis. |
|
|
Describe expected results of restriction analysis. |
- miniprep lanes have smear of degraded and partially digested chromosomal DNA, plasmid DNA, and RNA - at the front of the well are undissolved material and high-molec-weight DNA - cloud of low molec RNA at 100-200bp - uncut plasmids are seen |
|
|
If a plasmid has two ORI, is it still expressed? |
Yes but only one ORI is active at one time. |
|
|
Can a plasmid be made of odd-number of fragments after digestion? |
No. This is because BamHI sites only go with BamHI sites and same with HindIII sites. |
|
|
If a plasmid has two of the same fragment, can the fragment be adjacent to each other? |
No. Since they are the same, they have the same inverted repeats, which complement the entire fragment length. In DNA replication, this causes a hairpin loop, which is not viable. Thus, if there are duplicates, there need to be a different fragment in the middle. |
|
|
What is a simple recombinant? |
Only has the ampR and kanR fragments. |
|
|
If a third band of 784 pAMP is observed, what does it mean? |
- superplasmid where one of three fragment is repeated - simple recombinant and religated pAMP |
|
|
If third band of 2332bp (pKAN) is observed, what are some possible genotypes? |
- superplasmid with one of three fragments is repeated - simple recombinant and religated pKAN - simple recombinant and ligated 2332bp and ampR are together - religated pKAN and ligated 2332bp and ampR are together |
|
|
If all four fragments are seen, what is the interpretation? |
- superplasmid with all four fragments - pAMP and pKAN religation - simple recombinant and non-recombinant plasmids |
|
|
If four fragments are observed, with one pair (belonging to the same original plasmid) fainter than the other pair, what does it mean? |
- when bacteria is transformed with 2 different plasmids with related ORI; one plasmid is preferably replicated. Over time, one plasmid is lost, leading to fainter observation. |
|
|
In gel electrophoresis, where does the plasmid migrate? Pick two. a) + end b) - end c) anode d) cathode |
a) + end because DNA is negatively-charged; for this reason, also goes to anode |
|
|
What is the Ames test? |
A screening method for mutagens and thus, potential chemical carcinogens using bacteria. |
|
|
Who developed the Ames test? When? |
Bruce James in the 1970s |
|
|
Mammalian cell structure and enzymatic pathways differ from bacteria. Why does the Ames test still apply to humans? |
The chemical nature of DNA is common to all organisms. Mutations in bacteria are also seen in mammals. |
|
|
What is the test organism for Ames test? What are its properties? |
- histidine-negative, biotin-negative, auxotrophic Salmonella typhimurium - will not grow in a medium without histidine because it cannot make its own histidine - for growth to occur, a reversion to his+ must occur |
|
|
In mammals, where does toxification/detoxification (when a non-mutagen turns to a mutagen and vice-versa) occur? |
The liver - for this reason, we add S-9 (liver homogenate) to bacteria to contain metabolic enzymes found in mammals that cause (de)toxification. |
|
|
In the Ames test, what properties are shared by the mutation strains ? |
1) have no repair system normally found in wild-type bacteria; thus, mutation is not corrected. 2) defective lipopolysaccharide allowing for chemicals to penetrate more easily than in true bacteria |
|
|
What is the set up for the Ames test? |
- start with molten low melt agar, the test bacteria, 2-9 mix, trace of histidine and biotin (trace amounts allow bacteria to live for a few divisions; from here, we can get the bacteria that will mutate) - place disc impregnated with test chemical in the centre of plate - the chemical diffuses; most concentrated in the middle - incubate |
|
|
A colony is observed in the Ames test, what does this mean? |
The chemical is a mutagen and has caused a reversion from his- to his+, allowing for the colony to survive in environment with low histidine amount - compared to a negative control plate (no chemical added)... which may have a FEW colonies due to spontaneous reversion and not due to the chemical |
|
|
In this specific Ames test, what are some of the potential mutagens used? |
NaN3 (sodium azide), Scope, Listerine |
|
|
Why is there a cloudy lawn observed throughout the plate (not live colonies)? |
They are made of his- bacteria that survived initially when there were trace histidine but eventually died as histidine was depleted. |
|
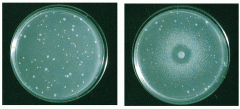
Describe this picture. |
Ames Test - in left, a negative control his- bacteria: the few colonies were due to spontaneous revertants - in right, the chemical kills the bacteria at very high concentration (zone of clearance); at slightly lower concentration, it acts as mutagen and causes his+ reversion, leading to many colonies; as we diffuse further, the concentration is so low that few colonies undergo reversion and perhaps, many of them were spontaneous revertants |
|
|
Quantitatively, what is the definition of a mutagen? |
- has min. twice the amount of reversion frequency than the negative control |
|
|
How does sodium azide cause mutations? |
- creates a point mutation |
|
|
What mechanism allows for transfer of chromosome of one bacterium to another? |
Conjugation |
|
|
If you mix 2 female strains, is it fertile? 2 males? Male and female? |
2 females: infertile; 2 males: poorly fertile; male and female: fertile |
|
|
What determines the maleness of a bacteria? |
The sex factor F ("fertility"); made of DNA and determines the function of male carrying it |
|
|
What properties does a male have, conferred by the F factor? |
1. Ability to form conjugal union with females. 2. One way transfer from male to female of genetic material.
|
|
|
What active role do females have in conjugation? |
No active role. They just receive genetic information. |
|
|
What are the two mutually exclusive states of the F factor in males? Is their high probability of alternating between the two? |
F+ sex factor is unassociated with the bacterial chromosome and is replicated independently. The Hfr state inserts into the chromosome and is replicated at the same time as the rest of the host chromosome. |
|
|
Which of the two states is more efficient when conjugating with females? |
Both are equally efficient. They only differ in how they are transferred. |
|
|
Name 3 properties of F+ male bacteria. |
1) On conjugation, sex factor transfer is efficient, converting the female to F+ male state. From here, sex factor spreads like a disease. 90% infection occurs in 1 hour. 2. Chromosome transfer is insignificant. Presence of actual chromosome is very low and is mostly due to presence of Hfr male in the F+ population. 3. Sex factor is eliminated from F+ male via acridine orange, reconverting to F- females. |
|
|
Name 5 properties of Hfr male bacteria (high frequency of recombinants). |
1. Hfr male bacteria arise from F+ when sex factor inserts into the bacterial chromosome. 2. Hfr male can transfer to F- female. F factor is nicked from chromosome's 0 (origin) or leading locus, moves to the female. Transfer is affected by orientation of sex factor. Thus, chromosome transfer may have different starting points or occur at different orientations. 3. Speed of transfer is temperature-dependent but constant at standard conditions. Transfer occurs in same order as in chromosome and intervals proportional to distance from origin. At 37C, takes 100 minutes for entire chromosome. They tend to randomly breakdown. Frequency is inverse exponentially proportional to distance (more frequent, the less distance). 4. Whole chromosome transfer is very rare, so recipient is still female. If you do find one with the entire chromosome, its most likely a Hfr male with the same chromosome as the donor. Functional part of sex factor is always transferred in the end. 5. No acridine orange effect.
1. Insert 2. Nick 3. T-sensitive; freq=distance 4. Rare whole chromosome transfer 5. No acridine orange |
|
|
What slows down transfer? |
Formation of conjugation tube. |
|
|
How do F' factors form? |
Hfr may revert to F+ due to reversion of recombination process but sometimes, the F+ may have part of the bacterial chromosome, making it an F'. |
|
|
What happens when F' is transferred to a F- female? |
-The recipient exhibits the F+ plasmid and part of the bacterial chromosome, forming an intermediate male. - low efficiency of transfer but same order
|
|
|
Describe zygotic induction. |
A Hfr male bacteria is lysogenic and carries a gene coding for inducible prophage. Since it is lysogenic, the prophage is not expressed. When this male mates with a non-lysogenic female (the zygote), it does not have the repressor to repress prophage expression. Consequently, the prophage is immediately expressed and the phage lyses the zygote. This does not work if female is also lysogenic |
|
|
How is zygotic induction useful? |
- no recombinants for genes distal to prophage since zygotes are lysed - prophages also act as chromosomal markers; since zygote is immediately lysed upon entrance of prophage, frequency of zygotic induction indicates frequency of transfer of gene (more lysing, the closer to the prophage). |
|
|
Describe episome transfer: spot making performed in this experiment. |
CSH23 are StrS wish contained F'lac+pro+; mated with CSH50 (lac-pro-strR) - plate into glucose minimal agar plate with streptomycin - if you plate only CSH23, they will die since they are StrS - if you plate only CSH50, there is some growth as streptomycin doesn't kill them. They first live off the glucose but once this is gone, they can't make proline and thus, will die. - if you plate both, CSH23 will still die. But some F'lac+pro+ may be transferred to CSH50. Thus, once glucose is diminished, they will continue to live. |
|
|
How do you determine efficiency of transfer? |
- by number of CSH50 recipient cells in population |
|
|
Liquid mating vs. Spot mating |
Liquid: Mix both strains. Dilute. Plate each dilution to minimal with streptomycin.
Spot mating: dividing one min. with strep. plate into three and spot CSH23 only, CSH50 only, and both. |
|
|
What do you do with liquid mating results? |
Count colonies and divide by mL of mating mixture. |
|
|
Stop and work on conjugation questions. |
OK. |
|
|
Is enzymatic machinery always turned on? |
No. It is only expressed when there is a present chemical signal. |
|
|
Does E. coli metabolize glucose at fast or slow rate? Under what conditions? |
Metabolizes fast at all conditions. |
|
|
What is the rate of lactose metabolism in E.coli grown in lactose and without lactose? |
When grown in lactose, lactose metabolism occurs at same rate as glucose because its enzyme machinery is always stimulated. When not grown in lactose, a lag occurs first before metabolism picks up because the enzyme machinery has to be activated. |
|
|
What enzyme in E.coli is responsible for lactose metabolism? |
B-galactosidase. Encoded by lacZ in lac operon of E.coli |
|
|
B-galactosidase breaks down lactose into what monosaccharides? |
Glucose and galactose |
|
|
Is B-galactosidase seen more in lactose or glucose-grown cells? |
Higher levels in lactose-grown. Rarely detectable in glucose grown. |
|
|
What is ONPG? |
-O-nitrophenyl-B-galactoside - structural analogue of lactose (thus, broken down by B-galactosidase) - when broken down, yields galactose and ONP (which has a yellow colour detected by spectrophotometry)
|
|
|
What is purpose of toluene in this experiment? |
unmasking agent that releases enzyme to the medium or buffer. |
|
|
What is IPTG? |
also a lactose analogue that inhibits lac repressor, strongly inducing B-galactosidase production - leads to breakdown of lactose and IPTG |
|
|
Describe the B-galactosidase essay experiment. |
1. Place toluene in 20 spectronic tubes. 2. To flask A, place lactose-grown E.coli cells in lactose solution. Upon transfer, treat this as time 0. 3. To flasks B,C,D, place glycerol-grown E.coli in lactose, glycerol, and IPTG solutions, respectively. 4. Incubate at 37C. 5. a)Every 15 minutes, take samples of A,B,C,D. b)Incubate in tubes toluene (step 1). c) Add ONPG. d)Water bath at 28C. e) Add Na2CO3 and water. 6) Read absorbance. |
|
|
What is purpose of Na2C03? |
Raises the pH so ONPG hydrolysis is stopped by B-Gal denaturation, so we get conditions only at that specific time. Also enhances colour. |
|
|
What is the expected result? |
- in flask A, no lag is expected. So, absorbance should be the same at all time as you're putting same amount of ONPG and this gets digested. - in flask B, there is lag because cells were not grown in lactose initially. However, since we now expose them to lactose, over time, there should be an increase in ONPG hydrolysis. - in flask C, no prior exposure to lactose. So, over time, we're not activating its machinery. ONPG may still get hydrolyzed but minimally. - in flask D, it is a lactose analogue and inducer of B-galactosidase. So, over time, hydrolysis should also increase. But, it decreases eventually as it not cleaved (no energy released to power cell) |
|
|
What wavelength of light does ONP best absorb light in? |
420nm. |
|
|
In one cycle, how many new phages can one infection create? |
100 or more |
|
|
When does cell growth stop after phage infection? |
When host is lysed and new phages are released to medium |
|
|
What happens when a lawn is infected with phage? |
- one cell is initially infected and is lysed, releasing phages to nearby cells, lysing them eventually as well - leads to a zone of clearance or plaque |
|
|
T/F: A plaque is caused by one phage and thus, the number of plaques represent the number of phages added to the lawn. |
True. It started from one phage which made more phages. |
|
|
How are titres represented? |
Plaque forming units/mL |
|
|
Typical phage solutions have titres of 10^8 to 10^11, making it difficult to count the plaques. How do we fix this? |
Dilute the solutions by serial dilution. |
|
|
Aside from the number of phages, what can we learn by observing the plaques? |
1) different types of phages/mutant phages as they make plaques of various appearances 2) make clones: each plaque has many phages of the same genotype |
|
|
What is the bacterial host used to grow bacteriophage in plaque assay called? |
indicator strain |
|
|
Describe the phage and its mutations used in the set up of the plaque assay experiment. |
You are given a bacteriophage λ stock. The phage has amber mutation (stop mutation) in the P gene, which is required for phage DNA replication and also T-sensitive mutation in repressor gene cI. - The latter ensures phage grows lytically at high T (when repressor is inactive). - due to amber mutation, phage will only grow in E.coli that have the gene that suppresses this amber mutation (sup), leading to DNA replication. - there are also some natural revertants (P+) in the stock which can grow even if E.coli has no sup. |
|
|
In plaque essay, describe the observation that E.coli strains MC4100 (supE+supF+ or sup0 or wild-type) had least plaques, YMC has intermediate, and C600 had the most plaques. |
- C600 is supE. This suppressor codes for a tRNA that reads the amber mutation codon and brings in an amino acid that is closest in property to what should have been there without the mutation. So, most of phages were expressed and performed lysis. - YMC (supF) coded for tRNA that brought less ideal amino acid - MC4100 did not have suppressors but plaques were due to natural revertants that didn't have amber |
|
|
Putting 0.1mL sample to 9.9mL diluent is what dilution? |
10^-1 dilution or 1/100 |
|
|
How do you calculate the concentration of phage in the original mixture? |
Cocn = #plaques/(dilution x Volume plated) |
|
|
Name examples of immunological assays |
agglutination, complement fixation, ELISA (enzyme-linked immunosorbant assay), immunodiffusion, RIA (radioimmunoassay), Western blot |
|
|
What are RFs? |
- rheumatoid factors - antibodies that recognize changed tertiary structure of IgG Fc fragments - found in human serum - used to track rheumatoid arthritis and auto-immune diseases |
|
|
Immunological assay can also track for bacteria such as Staphylococcus aureus. What diseases can be tracked through this method? |
meningitis, boils, toxic shock, food poisoning |
|
|
Do RF only recognize IgG? |
No. There are IgM, IgG, and IgA isotypes. |
|
|
Which arthritis has more favorable prognosis? |
Sero-negative is better than Sero-positive |
|
|
In agglutination to track RF, the latex particles are coated in what? |
- absorbed human gamma globulins |
|
|
How does agglutination occur? |
RF in test specimen reacts with coated material, leading to agglutination of inert latext particles |
|
|
What does RaPET stand for? What is it? |
Rapid Particle Enhanced Technology - uses the agglutination technique to detect RFs - positive serum causes globulins in RF to react with IgG coated latex particles |
|
|
Interpret the results for RAPET. |
positive means coarse agglutination in clear background while negative is smooth homogeneous suspension - agglutination means RF cocn of more than 20 IU/mL |
|
|
What is the titre of the serum? |
Reciprocal of highest dilution that exhibited a positive reaction (agglutionation) |
|
|
What is the limitation of RaPET? |
- if you leave the reaction for more than 2 minutes, drying effect may cause false positive |
|
|
What is BBL Staphyloslide Latext Test? |
differentiates staphylococci which have clumping factor and/or protein A (seen in S. aureus) from those without |
|
|
What is protein A? |
- protein found in surface of S. aureus that binds Fc portion of IghG |
|
|
What is clumping factor? |
Bound coagulase |
|
|
How does BBL Staphyloslide Latex Test work? |
blue latex particles coated with human fibrinogen and IgG - when exposed to clumping factor or protein A, cross-linking occurs leading to agglutination of latex |
|
|
Staphylococcus epidermidis gives positive results. T/F? |
False. |
|
|
Interpret results of BBL Staphyloslide Latex Test. |
- Positive: agglutination of blue latex is observed in 20 seconds AND negative control has no agglutination - Negative: no agglutination in 20 seconds - Uninterpretable: control shows agglutination |
|
|
Why does occasional granular or stringy rxn sometimes occur? How do we interpret this? |
Due to nature of test. Positive is there's a clearing of blue background, negative when there is no clearing. |
|
|
What are some limitations of BBL Staphyloslide Latex Test |
- other staphylococci give positive coagulase results - amount of expressed protein A depends on medium and conditions - MRSA have some clumping factor/protein A that may not be detected - S. saprphyticus found in urine gives positive results - autogluttination (partial clearing of blue background) due to overincubation (+36h); fix by retesting - E. coli can agglutinate but can be eliminated by Gram stain - salt leads to weaker rxns since colonies have harder time emuslifying |

