![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
150 Cards in this Set
- Front
- Back
|
Gametes
|
Only cells that reproduce through meiosis; derived from specialized germ cells in the ovaries and testes (ex: sperm and egg cells)
|
|
|
Mendelian Inheritance
|
Dominant and recessive allele inheritance theory. Does not hold true for genes on X and Y chromosomes
|
|
|
First Generation Cross
|
A cross between two true-breeding plants (ie: Mendel starting with two true-breeding pea plants and breeding them to create new plants)
|
|
|
Eukaryotic Cells
|
Cells with a nucleus (most of the time) that are part of a multicellular organism.
|
|
|
Prokaryotic Cells
|
Cells without a true nucleus that are single-celled organisms such as bacterium or a virus.
|
|
|
Microbiome
|
A population of microorganisms or microbes not visible to the naked eye (ie. streptococcus salivarius in the oral cavity and respiratory tract in humans. )
|
|
|
Cell
|
A membrane bound structure containing macromolecules.
|
|
|
Four Classes of Macromolecules
|
Nucleic Acids (DNA, RNA etc), Proteins (structure and metabolic activities), Polysaccharides (structural, sources of energy) and Phospholipids (primary components of cell membrane etc.)
|
|
|
Cell Membrane
|
A separation from an internal environment from the external world allowing for different chemical compositions for the two distinct environments.
|
|
|
Phospholipids
|
Make up the bilayer of the cell membrane, have a hydrophobic core and a hydrophilic surface. Made of a polar group, a phosphate group, glycerol, and two fatty acid chains.
|
|
|
Steriods
|
4-hrydrocarbon ring structures with a hydrophilic head and a hydrophobic tail (ex: cholesterol).
|
|
|
Lipid Micelles
|
Circles of phospholipids within the cell important for the absorption of fat-soluble vitamins; form spontaneously without the use of energy!
|
|
|
Fluidity of Cell Membrane
|
Unsaturated chains, higher temperature, and presence of cholesterol at low temperatures all contribute to this.
|
|
|
Movement of Cell Membrane
|
The phospholipid b. can move laterally but cannot easily flip from one side to another! (macromolecules move laterally)
|
|
|
Selective Permeability
|
The ability of cell membranes to control the traffic of substances into and out of the cell and its organelles.
|
|
|
Passive Diffusion
|
Small molecules/ions crossing membrane along a concentration gradient: High concentration --> Low concentration.
|
|
|
Facilitated Transport
|
Proteins embedded in the phospholipid bilayer spanning the whole width allowing for transport of large/polar molecules charges substances and lipid-insoluble molecules/ions to pass.
|
|
|
Fluid Mosaic Model
|
Membranes consist of proteins and carbs embedded in a fluid phospholipid bilayer (transmembrane).
|
|
|
Passive Transport
|
Facilitated diffusion and passive transport, from high to low concen.
|
|
|
Active Transport
|
Molecules movement against concen gradient using ATP
|
|
|
Osmosis
|
Movement of water across a membrane diffusing from less concentrated solution (hypotonic) to more concentrated solution (hypertonic)
|
|
|
Aquaporins
|
Transmembrane proteins allowing for rapid transport of of water (exclusively permeable to water), using osmosis-- is gradient dependent, uses no ATP
|
|
|
Isotonic
|
When there is no net movement of water across a gradient
|
|
|
Primary Active Transport
|
When there is direct expenditure of ATP for movement of molecules against a concen gradient.
|
|
|
Secondary Active Transport
|
When there is indirect expenditure of ATP for movement of molecules against a concen gradient.
|
|
|
Nucleic Acids
|
DNA-- deoxyribonucleic acid, contained in nucleoid/plasmids in prokaryotes, RNA-- ribonucleic acid
|
|
|
Chromosome
|
The organization of double-stranded DNA molecule in its association with proteins and RNAs; passed from parents to offspring in sexually reproducing organisms due to formation of sex cells/gametes transmitting genes form on gen to the next
|
|
|
Eukaryotic Chromosomes
|
Longer, larger linear organizations of DNA
|
|
|
Prokaryotic Chromosomes
|
Smaller, circular chromosomes that are supercoiled.
|
|
|
Supercoiling
|
Coiling in addition to the coil of the helical DNA to preserve the double helix and compact DNA into a small space.
|
|
|
Operon
|
A cluster of genes in a prokaryotic chromosome.
|
|
|
The Griffith's Experiment
|
S-Strain and R-strain of a virus injected into separate mice, S-strain is deadly but not when super-heated, r-strain not deadly no matter what BUT when injected together after both being super heated the mouse still died! Conclusion: living benign cells acquire info from dead virulent cells
|
|
|
Transformation
|
change in cell behaviour resulting from the incorporation of genetic material from outside the cell.
|
|
|
Avery and Colleagues Experiment (what is hereditary molecule)
|
When does transformation occur? Control of non-virulent and killed virulent bacteria with protein DNA and RNA, then had three more with one of those taken away (ie, RNA, DNA, or protein) Conclusion: DNA is the hereditary molecule
|
|
|
Rosalind Franklind
|
Created the x-ray diffraction image of the DNA molecule used to ID the helical nature and dimensions of DNA
|
|
|
DNA components
|
Phosphate group and 5-carbon sugar composing the sugar-phosphate backbone (3' to 5' polarity) and nitrogenous bases (A, T, C, G) within the helix connected with H-bonds!
|
|
|
Types of Nitrogenous Bases
|
Pyrimidines: Cytosine and Thymine
Purines: Adenine and Guanine |
|
|
Building DNA
|
DNA is built 3' to 5' via condensation reactions with the phosphate of the backbone being attached to the 3' hydroxyl with a phosphodiester linkage
|
|
|
Chargaff's Rules
|
DNA from any cell of all organisms should have a 1:1 ratio of purine to pyrimidine bases (C:G, A:T), also, G forms a triple H-bond with C and A forms a double with T.
|
|
|
From Franklin's X-Ray Diffraction Image
|
Diameter of DNA is 2nm, 10 base pairs (3.4nm) per one complete turn, and there are major and minor grooves along the outside of the helix
|
|
|
Three Categories of RNA
|
Messenger RNA, Ribosomal RNA, and Transfer RNA
|
|
|
Differences b/twn RNA and DNA
|
Backbone is ribose in RNA not deoxyribose, Uracil instead of Thymine (H instead of methyl CH3), and RNA is usually single-stranded.
|
|
|
Messenger RNA
|
Single-Stranded RNA molecule that is a copy of a gene that codes for a protein.
|
|
|
Transfer RNA (tRNA)
|
RNA that carries amino acids into the ribosome and matches the amino acid to the appropriate mRNA sequence
|
|
|
Ribosomal RNA (rRNA)
|
Form with proteins to create the functional ribosome complexes that perform translation of mRNAs into proteins
|
|
|
Tandem Repeats
|
Can be up to several thousand nucleotides in length and may be present next to each other in multiple identical or near identical copies
|
|
|
Simple-Sequence Repeats
|
Repeats as short as 2 nucleotides that can be repeated over and over throughout a DNA sequence stretch
|
|
|
Effect of Variations/Repeats in DNA
|
Will not always have an observed effect, esp in case of variations in non-coding regions-- some repetitions can be beneficial/harmful depending on the nature and location of the change (coding vs. non-coding regions etc.)
|
|
|
DNA Polymorphisms
|
One of Two or more alternative alleles at a chromosomal region (locus) that differs in either a single nucleotide base of has variable numbers of tandem repeats in a given pop. of individs.
|
|
|
Where are DNA Polymorphisms?
|
There is a large number of these changes across genomes of many organisms residing mostly in non-coding regions.
|
|
|
What do DNA Polymorphisms do?
|
Allow for assembly of high-density genetic maps and are often referred to as DNA-Markers as they are detectable using microarray analysis, PCR, and Southern blot to indicate where certain genes are/relatedness b/twn individs
|
|
|
SNPs
|
Single Nucleotide Polymorphisms, one of the most common types of genetic variation brought about by single nucleotide base changes/subs in a DNA sequence occurring in a significant potion of the pop. Often found scattered through the genome in coding and non-coding regions; 1:350 bp.
|
|
|
SNPs as DNA markers
|
SNPs found close to a particular gene can be used as a marker for that gene-- every time that gene is passed on from a parent to a child, that SNP is also passed
|
|
|
SNPs and Microarray Analysis
|
Oligonucleotides that match the common allele and all possible variant SNP alleles are attached to the glass of the microarray chip, millions of known single stranded oligonucleotides (containing a nucleotide base in the middle complementary to the SNP allele are attached too! Deliberate matches and mismatches. Fragments of flourescently labelled DNA are hybridized to the chip. The position of the oligonucleotide probes allow to match the flourescent pattern to which SNP an individ has.
|
|
|
VNTRs
|
Variable Number Tandem Repeats, can be linked to why species are different, are variations in short sequences of DNA repeated in tandem varying largely in number across individs
|
|
|
Tandem Repeats
|
Patterns of one or more nucleotides that are repeated directly adjacent to each other found in various lengths b/twn different individs across pops. ID'd using PCR and gel electrophoresis (TR sites targeted and amplified with sequence specific primers targeting the flanking regions of the VRs resulting in DNA fragments that can be separated and run through a gel.
|
|
|
DNA Fingerprinting
|
Used for IDing individs based on their respective DNA profiles, utilizes variable number of tandem repeats (as VNTR loci are similar but variable enough so that 2 people will most likely not be the same) genetic family relationships or crime scene evidence analyzed.
|
|
|
Variation in Non-Coding regions of Human Genome
|
Most variations occur here and have no known effect-- are detectable through many molecular techniques but are often called "silent variations"
|
|
|
Variation in Coding/Regulatory Regions of Human Genome
|
Variations that can be harmful as they will result in the production of altered gene product leading to possible detrimental effects in the organism (ex: sickle cell anemia)
|
|
|
Genotype
|
Representation of the pair of alleles carried by a person
|
|
|
Phenotype
|
Cell/body's interpretation of the genotype
|
|
|
Cellular Phenotype of Sickle Cells
|
Variation of the Oxygen binding hemoglobin protein assuming a sickled shape distinct from the regular biconcave shape
|
|
|
Physiological Phenotype of Sickle Cell
|
Seen at diff. levels of an organism: oxygen not carried efficiently around the body, sickle cells can block fine capillaries of circ system leading to anemia and acute pain throughout the body
|
|
|
Gene for Beta-globin protein
|
Gene that lies on an autosome (chromosome 11) meaning every person carries 2 alleles for the gene (ma and pa), most people have HbA alleles with sequences coding for the functional B-globin protein (homo for HbA).
|
|
|
HbS
|
Sickle cell allele brought about by a single nucleotide polymorphism occuring in protein coding sequence of the B-hemoglobin gene (coding for valine instead of glutamine)
|
|
|
Having 2 HbS Alleles
|
Homo, caused by simple mutation leading to an amino acid sub of glu --> val, altering the 3D (tertiary) structure of the final hemoglobin protein. Biochemical change in B-hemoglobin protein resulting in aggregation of long rod-like structures within red-blood cells (sickle shape).
|
|
|
Hetero HbA/HbS
|
Will still develop sickle cell trait; some sickle under certain conditions the rest normal therefore the person exhibits no symptoms: production of enough normal hemo to overcome the effect of the abnormal
|
|
|
Where is Sickle Cell Found?
|
This disease is found in many pops. with different alleles/haplotypes and emerges independently across various populations (in highest prevalence 5 distinct B-globin found across patients correlating with regional distribution of each distinct sickle cell SNP-- one haplotype distributed across nations or many within one).
|
|
|
CNVs
|
Copy Number Variations: contribute to genetic differences, some occur in non-coding regions or are present as many tandem repeats of a coding region along a chromosome resulting in region of genome being duplicated/deleted
|
|
|
IDing CNVs
|
Copy Number Variations are ID's based on relative flourescence intensities detected during DNA microarray analysis resulting in greater number of copies of chromosome sequence indicated by higher flourescence outputs on the microarray chip
|
|
|
Effect of gene Copy # on Phenotype
|
Gene duplications usually found adjacent to each other on the chromosome and the more there are, the more gene product is produced (as in AMY1 and salivary amylase)
|
|
|
Sexual vs. Asexual Reproduction
|
Sexually produced offspring are not identical copies of their parents whereas asexually produced are usually
|
|
|
Diploid Offspring
|
Offspring with unique combo of genes inherited from both parents (production of gametes in both parents from replication of parental sex cell precursors), reduced diploid to haploid chromosome number in final gametes because of 2 successive rounds of meiotic cell division.
|
|
|
Human Sperm
|
There is production of 4 haploid sperm cells from one sex cell precursor cell (found in testes)
|
|
|
Human Eggs
|
Produced in similar manner to sperm but there is often one large egg cell produced with higher cytoplasmic volume and 3 non-gametic polar bodies which serve the role of reducing the chromosomal content of the egg.
|
|
|
Diploid/Haploid Number
|
Each sexually reproducing organism has a characteristic diploid/haploid number (ie humans 46/23)
|
|
|
Meiosis
|
The process that leads to the reduction of chromosome number in gametes allowing for recombination of parental homologous chromosomes therefore the production of unique and variable gametes
|
|
|
Stages of Meiosis
|
Interphase, Prophase 1, Metaphase 1, Anaphase 1, Telophase 1, (Cytokinesis 1), Prophase II, Metaphase II, Anaphase II, Telophase II, (Cytokinesis II).
|
|
|
Interphase (meiosis)
|
Phase allowing for duplication of chromosomes to occur; subsequently followed by 2 consecutive cell divisions.
|
|
|
Prophase I
|
Phase characterized by chromosome condensation and synapsis (pairing of and physical connection) of homologous chromosomes along their length, facilitated by synaptonemal complex forming b/twn homologous chromosomes holding them together during synapsis. Crossing over, chromosome duplication and movement, spindle formation, nuclear envelope breakdown.
|
|
|
Homologous Chromosomes
|
Individ chromosomes inherited from each parent (may have diff versions/alleles of the same genes at the specific loci for specific genes)
|
|
|
Daughter Cells
|
The final product of meiosis: 4 cells all genetically different from each other and their parental cell.
|
|
|
Crossing Over
|
Occurs b/twn homologous chromosome pairs resulting in a physical breakage and reunion b/twn non-sister chromatids producing chiasmata along the chromosomes- is random and can occur anywhere along the length of the paired homo chromo allowing for the production of recombinant chromatids
|
|
|
Chiasma/Chiasmata
|
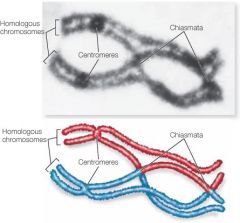
X-shaped regions on paired chromosomes where crossing over occurs.
|
|
|
Recombinant Chromatids
|
Chromatids carrying some paternal and some maternal segments of genetic info contributing to very distinct gametes which results in an increase in genetic diversity (left over after crossing over)
|
|
|
Bivalent Unit
|
Consists of a pair of synapsed chromosomes which thusly form a four-stranded structure with chromatids attached to different centromeres (non-sister chromatids)
|
|
|
Sister Chromatids
|
Duplications of the same chromosome and therefore genetically identical to each other (present in the beginning of meiosis and in mitosis)
|
|
|
Non-Sister Chromatids
|
Replicas of different chromosomes which are genetically similar but not identical to each other b/c of one pair of homologous chromosomes is maternal and the other is paternal in origin.
|
|
|
Metaphase I
|
Pairs of homo chromo bivalents become randomly arranged relative to each other at the metaphase plate by the microtubules of the spindle apparatus increasing the genetic diversity of gametes.
|
|
|
Anaphase I
|
The proteins that hold the homo chromo pairs together (the synaptonemal complex) will break down allowing for separation of the homologues making up each bivalent o the opposite poles of the cell
|
|
|
Meiosis different from Mitosis
|
Left with 4 haploid daughter cells all genetically different from each other and the parent, homo chromos separate and sister chromatids don't
|
|
|
Telophase I
|
End of Meiosis I, each half of cell has a complete haploid set of the duplicated chromosomes with each chromo consisting of a pair of recombinant sister chromatids, the chromosomes will slightly uncoil in this phase and the nuclear envelope will reform
|
|
|
Meiosis I
|
Reductional Cell division (reduction in chromosome #, Ex: in human gamete formation, the parent diploid, 46 chromo, has divided into 2 haploid, with 23 chromos).
|
|
|
Meiosis II
|
Equational Division, no chromosome duplication occurs b/twn I and II therefore there is no second interphase!!
|
|
|
Prophase II
|
The nuclear envelope breaks down, the spindle apparatus forms, and the chromosomes condense.
|
|
|
Metaphase II
|
The chromosomes (each consisting of 2 sister chromatids) are positioned at the metaphase plate, 2 sister chromatids are genetically identical.
|
|
|
Anaphase II
|
Proteins that hold sister chromatids together at centromere are broken down allowing the chromatids to separate and move to opposite poles of the cell as individ chromosomes.
|
|
|
Telophase II
|
Marks end of meiosis II, reformation of nuclear envelope and chromosomal decondensing. (is followed by cytokinesis)
|
|
|
Nondisjunction
|
Cases where members of a pair of homo chromos in meiosis I or sister chromatids in meiosis II fail to separate resulting in some gametes with extra chromos and some with missing-- can lead to detrimental and sometimes lethal effects in generated offspring (can occur in mitosis, often seen in cancer cells).
|
|
|
Heredity Concept early 1800s
|
Based on a blending of hypothesis in which it was believed that genetic material from both parents is mixed to produce the visible traits observed in offspring.
|
|
|
Gregor Mendel
|
A monk who ID's and documented 2 laws that explain the basic principles of inheritance based on the traits observed in pea plants; paved the way for modern transmission genetics
|
|
|
central dogma
|
The process of copying and interpreting genes into proteins first proposed by Francis Crick in the late 1950s
|
|
|
Transcription in prokaryotes
|
Initiated when sigma factor binds with RNA Polymerase II; RNA polymerase will move along the template strand in a 3’ to 5’ direction until a nucleotide sequence on the template strand known as the terminator will stop transcription and lead to release of the RNA transcript (end of a specific gene)
|
|
|
Promoter Regions
|
Regions indicating the transcriptional starting point where RNA synthesis actually begins found upstream to gene of interest (5’)
|
|
|
Prokaryotic DNA
|
DNA not packaged into chromosomes, but instead sits within a non-membrane bound nucleoid regionprokaryote genome is smaller and has fewer genes than eukaryotes
|
|
|
consensus sequences
|
Regions before a gene in prokaryotic DNA that enhance the rate of transcription; Ex: TATAAT in prokaryotes; also serves as part of the promoter region.
|
|
|
sigma factors
|
Proteins in prokaryotes which facilitate the binding to the promoter region of the DNA, recruited by RNA polymerase to bind to its core to create a holoenzyme. Also identify certain promoter regions to the RNA polymerase.
|
|
|
Holoenzyme
|
An RNA polymerase core enzyme bound to a sigma subunit capable of binding to and unwinding the double-stranded DNA helix to allow transcription to occur
|
|
|
transcription bubble
|
RNA polymerase separates the two strands of the DNA double helix allowing ribonucleotides to enter the RNA polymerase, and assemble in a complementary fashion on the DNA template strand within this!
|
|
|
Prokaryotic Terminator sequences
|
Rho-independent and Rho-dependent
|
|
|
Phosphodiester Bond Formation
|
phosphate bond energy of the incoming ribonucleoside triphosphate is used to drive the high energy reaction process that is required to create this bond. The release and cleavage of the pyrophosphate (or phosphate-phosphate) group during this bond formation renders RNA elongation an irreversible process!
|
|
|
Rho-independent
|
Terminator sequences consisting of inverted nucleotide repeat sequences. They are transcribed and then fold back on themselves to form what is referred to as a G-C rich hairpin loop along that same mRNA strand: pauses the RNA polymerase and leads to the release of the mRNA transcript.
|
|
|
inverted nucleotide repeat sequences
|
Sequences of nucleotides that are usually followed downstream by a specific reverse complement.
|
|
|
Rho-dependent
|
Termination that uses a specific prokaryotic protein or Rho factor, which can bind to and subsequently utilize ATP energy to move along the formed RNA transcript while unwinding it from the DNA template. The Rho factor is able to destabilize the interaction between RNA and the DNA template, leading to the release of the transcript and the transcription complex
|
|
|
general transcription factors
|
A number of specific proteins in eukaryotes that are required to mediate the binding of RNA polymerase to a promoter and to initiate transcription
|
|
|
RNA Polymerase I
|
Polymerase that transcribes the genes for the ribosomal RNAs (rRNAs) (transcribe-structural, non-coding RNAs) (terminated via a specific eukaryotic termination factor in a similar manner as prokaryotic rho-dependent termination)
|
|
|
RNA Polymerase II
|
Polymerase that transcribes the messenger RNAs. Termination depends on a poly(A)-dependent mechanism; termination of transcription is coupled with mRNA maturation where the 3’ end of the transcript is modified by polyadenylation and modification of the 3’ mRNA transcript is coupled with the termination of transcription.
|
|
|
RNA Polymerase III
|
Polymerase that transcribes structural, non-coding RNAs; transcribes the genes for the transfer RNAs (tRNAs), as well as other small regulatory RNA molecules (have transcription terminated after transcription of a termination sequence in a mechanism that resembles the rho-independent termination in prokaryotes)
|
|
|
core promoter consensus sequences
|
Sequences required to set up a transcription initiation complex, specific for the particular RNA polymerase and gene being transcribed.
|
|
|
mature mRNA
|
Changes to ensure stability of the mRNA molecule, ensure the export of the mRNA from the nucleus, help protect against ribonuclease enzymes that target phosphodiester bonds, and help with attachment of the ribosome and initiation of translation once the mRNA reaches the cytoplasm of the cell
|
|
|
5' Cap
|
Capping of the 5’ end of an mRNA transcript involves the attachment of a modified guanosine ( 7-methylguanosine since it also has a methyl group (-CH3) that is attached to the 7th position in guanine) to the mRNA through an unusual 5’ to 5’ triphosphate linkage: terminal 5’ phosphate is removed from the mRNA molecule by a phosphatase enzyme, and another enzyme, guanosyl transferase enzyme, catalyzes the attachment of the 7-methylguanosine 5’ cap
|
|
|
Poly-A Tail
|
Additional adenine nucleotides are added to the 3’ end of the mRNA transcript following recognition of a polyadenylation signal sequence (AATAAA) that is transcribed from the DNA template strand near the end of the gene sequence
|
|
|
polyadenylation
|
The process of the mRNA being cleaved and a poly (A) polymerase enzyme is able to add between 150-200 adenine nucleotide bases to the 3’ end of the RNA transcript and is coordinated with termination of transcription.
|
|
|
Exons
|
The sequences of the mRNA that are necessary for coding the sequence of amino acids in the protein that need to be joined or spliced together prior to translation through a process called RNA splicing
|
|
|
Introns
|
The intervening sequences that are not used for amino acid coding that need to be removed or excised from the mRNA
|
|
|
pedigree
|
family history for a particular trait that allows for a visual representation of thesegregation of the specific trait of interest and males in the family are typicallyrepresented by squares and females arerepresented by circles and the order of birth is identifiedfrom left to right.
|
|
|
Recombination b/twn X and Y chromosomes?
|
NO! This will not occur because they are largely nonhomologous,meaning almost none of the genesin the X chromosome have counterparts in the Ychromosome. only smallregions at the tips of the X and Y chromosomesthat allow for these chromosomes to pair andsegregate like homologous chromosomes duringmeiosis
|
|
|
Human Y chromosome
|
78genes on this chromosome that code for about 25proteins (and half of these genes help determinesex) --leads to the developmentof the male phenotype
|
|
|
Human X chromosome
|
Has 1100 genes with many functions that areunrelated to sex determination. p arm (or short arm) and onthe q arm (or long arm)
|
|
|
sex-linked gene
|
A gene that islocated on either sex chromosome (as expression of the phenotype may depend on thesex of the individual)
|
|
|
Ishihara colourtest
|
one of the most common ways to test forred/green colour-blindness; consists ofa circle containing different coloured dots with aninternal pattern of dots that forms a numbershape (such as the number 5 seen above) that isclearly visible to individuals with normal colourvision and invisible or difficult to see by colourblindindividuals
|
|
|
hemizygousindividual
|
only having one locus for an allele (ie male having only one X-chromosomes for certain alleles such as colour-blindness). In the hemizygousstate, the rule of dominance and recessivenessno longer apply
|
|
|
Passing on Colour-Blindness Allele
|
Given the principles of Mendelian inheritance,there is a 50% chance of passing on the allelethat determines colour-blindness.
|
|
|
haemophilia
|
blood-clottingdisorder; X-linked recessivetrait that results from a mutation in a gene thatencodes for a necessary protein that is requiredfor blood clotting
|
|
|
Mendel’s second law of inheritance
|
Law stating that two genes sort independently during gameteformation
|
|
|
linked genes
|
Genes that arepositioned close together on the samechromosome (whether they are on homologousautosomes or X chromosomes) They tend to be inheritedtogether and do not segregate independently. ** If the linked genesare far enough apart, recombinant chromatidsthat carry alternate allele combinations aregenerated during crossover events.
|
|
|
Recombination frequency
|
toolthat can be used to determine the distancebetween genes along the same chromosome since the recombinationbetween linked genes is dependent on thedistance between them, genes that are closertogether show less recombination frequency thangenes that are located far apart from each other.
|
|
|
High-density linkage maps/ SNP-linkage maps
|
identify genetic loci that are merely a fewthousand base pairs apart and can thenbe used to map human genes that determinevarious characteristics (looking at SNPs as gene markers)
|
|
|
GWAS
|
A Genome-Wide Association Study looks across the entire SNP-linkage map for anassociation between a particular phenotype and amapped SNP
|
|
|
HMGA2 gene
|
Association between height and a particularmarker allele, researchers were able to identify this gene that contributes to increased height inhumans. contributes toless than 1 cm variation in height. Individualswith two “C” alleles of HMGA2 are 0.8cm tallerthan people with two “T” alleles. Individualsheterozygous for the “C” and “T” alleles are only0.4cm taller than people with the two “C” alleles.
|
|
|
cellularproteomes
|
Driven by different transcriptionalprograms that are able to direct different cells toengage in specific functions in association withother cells or even within tissues.
|
|
|
ABO blood typing system
|
A and B dominant, O recessive! A or B agglutinogens thatare expressed on the cell surface and the O alleleencodes an inactive glycosyltransferase. AB blood type has several SNPpolymorphisms that leads to the formation ofslightly different transferases.
|
|
|
A blood type
|
a classification of blood based on the presence or absence of specific inherited cell surface proteins, or even enzymes that catalyze the synthesis of cell surface carbohydrates or glycolipids in different individuals.
|
|
|
ABO blood typing system
|
The ABO locus has three main alleles- A,B and O. The A and B alleles code for a specificglycosyltransferase enzyme that catalyzes theformation of specific A or B agglutinogens thatare expressed on the cell surface and the O alleleencodes an inactive glycosyltransferase. Inaddition the AB blood type has several SNPpolymorphisms that leads to the formation ofslightly different transferases. Since these allelesare genetically inherited from our parents (onefrom each parent), the two allele copies of theABO gene lead to our specific blood type.
|
|
|
immunity to HIV infection
|
This mutation is a 32 base pair deletion within theCCR5 gene. This type of deletion is not in asequence of three, and thus shifts the readingframe such that a stop codon can terminatetranslation early and the result is the productionof a partial and inactive CCR5 protein
|
|
|
variation in microbiome populations
|
does notcorrelate with ancestry, but instead is associatedwith recent dietary patterns. Diets high in animalproteins and fats are associated with a differentmicrobiome population than diets high in plantbasedsources
|
|
|
Mendel's experiments Terminology
|
P/parental generation
filial1 or F1 generation: the produced offspring |
|
|
Mendel’s first generation cross
|
gave evidenceagainst the blended model of inheritance. All theF1 offspring had a yellow seed colour.
|
|
|
Steps of RNA splicing
|
1. RNA in the spliceosome is able to recognize and form complementary base pairing with nucleotides at the designate splice sites
2. It then catalyzes a reaction that allows for a specific hydroxyl group on a nucleotide in the branch site to effectively “attack” and form a new phosphodiester bond with a nucleotide at the donor site 3. the cut 5’ end of the intron to then form a lariat intermediate which forms a loop within the intron 4. the 3’ hydroxyl group at the cut end of the 5’ exon at the donor site is able to “attack” the phosphodiester bond at the 3’ acceptor site, forming a new phosphodiester bond between the 3’ end of the upstream exon and the 5’ end of a downstream exon. 5. allows for the release of the intron and the splicing of the exons together in much like a cut and paste mechanism 6. The excised intron lariat is subsequently broken down into individual nucleotides that can be recycled by the cell. |
|
|
nuclear pore complexes
|
export of mRNA from the nuclear compartment to the cytoplasm is facilitated by these
protein-lined channels that pass through both membranes of the nuclear envelope (both the outer and inner membrane)In addition to the transport of RNA from the nucleus to the cytoplasm, nuclear pores can also transport proteins, carbohydrates and important signalling molecules into the nucleus. |

