![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
125 Cards in this Set
- Front
- Back
|
Explain the interaction between antigens and immunoglobulins
|
Antigens are picked up by receptors (immunoglobulins) on the surface of B lymphocytes
|
|
|
Describe the constant and variable regions on the immunoglobulin
|
- VH and VL have consistency
- CH and CL and for effector and signalling |
|
|
Explain the Dreyer and Bennett (1965) concept
|
- Ab protein is encoded more than one gene
- each receptor chain cannot be encoded by just one gene - more genes than in genome - V regions encoded by assembled gene segments - |
|
|
How could this large Ab repertoire
be generated? |
- cannot be encoded in the entire genome
- would require more genes than in the genome - instead the variable regions are encoded by gene segments |
|
|
What is gene rearrangement and what does it do
|
- two or three types of gene segments which are in multiple copies in the genome
- occurs at random |
|
|
Ab diversity is the marriage of two theories - what are they?
|
1.germline theory
2. somatic diversification |
|
|
What is the evidence for somatic identification?
|
Southern blot: DNA from non- lymphoid tissues
and CLL (B cell tumor) cut with restriction endonuclease Probed with V-region and C-region probes In B cells DNA is rearranged so that V and C regions are found on the same DNA fragment Susumu Tonegawa 1987 Nobel prize in medicine Somatic recombination events were able to take a limited amount of genetic material and create almost innumerable permutations, encoding a vast antigen receptor repertoire |
|
|
How are VH and VL regions encoded?
|
- by multiple germline gene families
- Complete V regions genes are generated by breaking and re-joining gene segments that are selected at random from gene families |
|
|
Complete V regions genes are generated by
|
breaking and re-joining gene segments that are
selected at random from gene families |
|
|
Explain Joining of gene segments
|
- not precise
- Nucleotides are added or subtracted at joining sites in a random manner |
|
|
how large is the antibody repertoire?
|
10^11
|
|
|
Dscribe the germline theory
|
that there was a separate gene for each individual immunoglobulin
|
|
|
Describe somatic diversitification therory
|
Ab repertoire was generated from limited number of V region sequences that undergo alteration with B cells. This involves hypermutation and cloning - somatic recombination is different from meitotic recombination
|
|
|
How many gene segments encode the V light chain domain?
|
- two separate
- V gene 95-101 aa - J gene segment (joining) segment 13 aa |
|
|
Describe the variab;e V gene segment
|
- encodes the greater part of the Variable Light chain
- 95 - 101 amino acids |
|
|
Describe the joining or J gene segment
|
- encodes the remainder of the variable light chain domain after V segment
- up yo 13 amino acids |
|
|
Describe what occurs in rearrangements that produce a complete immunoglobulin light chain gene
|
- joining of the V and J chains creates an exon
- exon encodes the entire light chain V region - V gene segments are located far away from the C region - J and C are close - when V and J combine - it is also with the C region - to make light chain mRNA the V region exon is joined to the C region sequence by RNA splicing after transcription |
|
|
Describe what occurs in rearrangements that produce a complete immunoglobulin heavy chain gene
|
- encoded in three segments
- there is a D gene segment - D (diversity) segment in addition to V and J - D lies between the V and J segments - occurs in two separate stages 1. D + J 2. V + DJ = complete exon - RNA splicing joins the assembled V region sequence to neighboring C region gene |
|
|
Describe random selection in V segments -
|
multiple copies of all gene segments in germline DNA and the random selection og just one segment of each type that makes possible the great diversity of V regions among immunoglobulins
|
|
|
Define pseudogenes are their role
|
non functional gene segments. Accrued permutations have rendered the gene segment as unable to be encoded as a functional protein
|
|
|
What are the three genetic loci?
|
- gamma light chain locus
- kappa light chain locus - heavy chain locus |
|
|
Describe the constituents of complete light chain V genes
|
Complete light chain V genes consist of one V gene and one J gene selected at random from the V
and J gene families |
|
|
Complete heavy chain V genes consist of ...
|
1 V, 1 D + 1 J gene selected from the V,D and J
gene families |
|
|
How many variable segments in the kappa light chain are available?
|
40
|
|
|
How many J segments are available for the light gamma chain?
|
5
|
|
|
How many variable segments are available for the gamma light chain?
|
30
|
|
|
How many joining segments are available for the gamma light chain?
|
4
|
|
|
How many variable, diversity and joining sements are available for the heavy chain?
|
40,25, 6
|
|
|
What is the First mechanism for generation
of diversity? |
**
|
|
|
Describe Sequence of stages / cell types of B cell development
|
1. stem cell
2. early pro B cell 3. late pro-B cell 4. Large pre-B cell 5. small pre-B cell 6. immature b cell 7. mature b cell |
|
|
Describe the H chain genes in B cell development
|
- D-J rearranging occurs at early pro B cell stage
- V -DJ rearranging occurs at late pro B cell |
|
|
What is the stage of b cell development where we see H chain gene rearrangement
|
early and late pro b cells (2,3)
|
|
|
What is the stage of B cell development where we see L chain genes rearrange?
|
V-J rearrange at small pre-B cell and are finished rearranging at immature B cell
|
|
|
When do we see diversity in L chains in B cell development?
|
immature b cells
|
|
|
Describe when we see surface immunoglobulins in b cell development?
|
1. in large pre-B cell the mu chain transiently at surface as part of the pre-B cell receptor, but mainly intracellular
2. small pre-B cell we see an intracellular mu chain 3. IgM is expressed on the surface of immature B cells 4. IgD is now with IgM on mature B cells |
|
|
Which locus arranges first in B cell development?
|
heavy chain
|
|
|
Summarise heavy chain rearrangement
|
Heavy chains: 2 chromosomes, 2 attempts to generate a functional heavy VDJ gene
|
|
|
Which stage of B cell undergoes +++ proliferation and how does this relate to gene arrangement?
|
-Large pre-B cells undergo multiple rounds of division. Increases the number of cells that can
attempt light chain gene rearrangement |
|
|
Describe which loci where light chain rearrangement can occur
|
-Light chains: rearrangement can occur at both κ and λ loci, on both chromosomes
|
|
|
Define recombination signal sequences
|
non coding DNA sequences found adjacent where recombination takes place. RSSs are composed of
- heptamer (7n) - nonamer (9n) and these are separated by a spacer which is either 12bp or 23bp long |
|
|
What is the heptamer code>
|
5'CACAGTG3'
|
|
|
What is the purpose of RSSs?
|
Conserved heptamers and nonamers flank VDJ genes that are to be joined, therefore facilitating DNA rearrangement
|
|
|
What is the nonaner code>
|
ACAAAAACC
|
|
|
How many bp long is a spacer
|
either 12 or 23,
|
|
|
Why can V and J not join directly?
|
Due to the 12/23 rule. for heavy chain the d hene segment can be joined to the j segment but the v segment cannoit be joined directly. This is due to the fact that both V and J segments have 23 bp for spacers.
|
|
|
Which spacer is one turn of the DNA double helix?
|
n 12 nucleotides = 1 turn of a DNA double helix
|
|
|
Explain the 12/23 rule
|
A gene sequence flanked by a RSS with a 12bp spacer can only join with a gene sequence flanked by a 23bp spacer
|
|
|
Give an example of the 12/23 rule
|
Thus in heavy chain gene rearrangement:
VH cannot join to JH segments directly as both have 23 bp spacer |
|
|
What do 12/23 spacers do?
|
n 12/23 spacers align gene segments to be joined
|
|
|
What is the role of RSS heptamers and nonamers ?
|
RSS heptamers and nonamers serve as binding sites for enzymes that cleave and rejoin DNA fragments
|
|
|
What are the two different types of recmbinations?
|
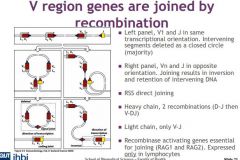
1. SAME ORIENTATION two segments undergoing rearrangement are in the same transcriptional orientation in the chromosome -- looped and deleted
2. OPPOSITE TX: gene segments are initiatlly orientated in opposite tx directions -- inversion and integration |
|
|
Describe the process of recombination when gene segments are in the same direction of transcription
|
recombination occurs at the end of heptamer sequences in the RSSs, creating the so called signal joint, and releasing the intevening DNA in the form of a closed circle.
|
|
|
Describe the process of recombination when gene segments are in the opposite transcriptional direction
|
gene segments are in opposite tx directions in this case, it requires more complex looping of the DNA. The ends of the heptamer sequences are joined, but this inverts the intervening DNA - which will now be integrated in the rearranged chromosome and not spliced out.
|
|
|
What is the difference between recombination in the light chain vs recombination in the heavy chain?
|
Heavy chain, 2 recombinations (D-J then
V-DJ) Light chain, 1 recombination - only VJ |
|
|
Describe role of RAG1 and 2
|
Recombinase activating genes essential
for joining (RAG1 and RAG2). Expressed only in lymphocytes |
|
|
How are RSSs brought together?
|
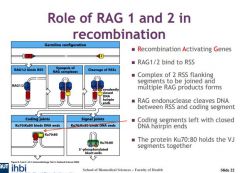
interaction between enzymes which detect the length of spacers. DNA molecule is broken in two places at the heptamer - where the heptamers are head to head and form a signla joint. The signal joint is then spliced in to a circular extrachromosomal DNA which is lost in the next cell division
|
|
|
Define coding joint
|
V and J gene segments remain on the chromosome and join to form the coding joint.
|
|
|
What happens to the signal joint in inversion?
|
It is retained in the chromosomal DNA
|
|
|
What does RAG stand for>
|
Recombination activating genes
|
|
|
What do RAG1:2 do?
|
RAG endonuclease cleaves DNA
between RSS and coding segment |
|
|
What is the function of the Ku70:80 protein?
|
The protein Ku70:80 holds the VJ
segments together |
|
|
Describe the steps in RAG dependent V(D)J rearragnement
|
1. RAG1:2 binds RSS
2. Synapsis of RSS where the two complexes are brought together 3. cleavage of RSS - endonuclease splits DNA in to coding and signal joints. 4. Both Signal and Coding joints are bound by Ku70:80 5. Coding: DNA-PL:Artemis opens hairpin Signalling: DNA ligase IV:XRCC4 ligases DNA ends 6. Coding: TdT processes DNA ends signalling: circular precise signal joint 7. DNA ligase IV:XRCC4 ligates DNA ends |
|
|
What chromosome is the light chain gamma locus located?
|
chromosome 22
|
|
|
Which chromosome is the kappa light chain locus located?
|
chromosome 2
|
|
|
Which chromosome is the heavy chain locus located?
|
chromosome 14
|
|
|
What is the role of DNA-PK
|
- Ku70:Ku80, a heterodimer which forms a ring around the DNA and associates wit a protein kinase catalytic subunit DNAPKcs to form DNA-PK
- DNA-dependent protein kinase (DNAPK) involved in artemis recruitment and eventual DNA repair |
|
|
What is the role of Artemis
|
- Protein with nuclease activity which acts with DNA dependent kinase,
- Artemis (a nuclease) binds to complex and opens DNA hairpins |
|
|
What is the role of Ku 70:80
|
DNA modifying protein involved with the repair of ds DNA breaks and modification of the ends of broken DNA strands
|
|
|
What is the role of DNA ligase IV
|
enzyme that joins together the ends of ds DNA broke during gene rearragnements that generate functional genes for Ig and TCR
|
|
|
What is the role of XRCC4?
|
DNA repair protein which forms a complex with DNA ligase IV
|
|
|
What is the role of RAG-1 and RAG-2
|
Recombinase proteins which are essential for Ig and T cell receptor gene rearrangement
|
|
|
What is TdT?
|
Terminal deoxynucleotidyl transferase (TdT) processes DNA ends
- adds randomly adds nucleotides to both strands - nonproductive rearrangements, many cells die at this stage |
|
|
Describe how ssDNA overhangs come about
|
- Artemis cleavage of hairpin results in short ssDNA overhang
- ssDNA repair enzymes fill in these ssDNA overhangs (TdT) randomly adds nucleotides to the ssDNA overhang (up to 20) |
|
|
light chains
|
V,J, and C
|
|
|
heavy chains
|
V,J,D
|
|
|
Light chains are made of:
A) one V and one J segment in the variable region plus a constant region that is common in all light chains B) one V, one D, and one J segment in the variable region plus a constant region that is common in all light chains C) one V and one J segment in the variable region plus one of two possible different constant region segments D) one V and one J in the variable region and no constant region |
one V and one J segment in the variable region plus one of two possible different constant region segments
|
|
|
2. Heavy chains
A) all have the same constant region B) have one of two possible constant regions C) have one of five possible constant regions D) have one of eight possible constant regions |
have one of five possible constant regions
|
|
|
The V, J, and D segments
A) must be a matching set B) are each selected randomly C) are present in the constant region of the heavy chain D) are RNA sequences |
??are each selected randomly
|
|
|
Human cells have separate genes for each antibody molecule.
A) True B) False |
false
|
|
|
5. The m (μ) gene codes for the constant region of the light chain in IgM antibody.
A) True B) False |
FALSE
- mu is a gene segment - mu will be a isotype of the heavy chain of an IgM |
|
|
6. The variable region of the heavy chain and the variable region of the light chain in a given antibody
molecule: A) are identical to each other in primary amino acid sequence. B) are each encoded by single germ-line gene segments. C) in combination with one another, define the specificity of the antibody for an antigen. D) contain the hypervariable and constant region portions of an antibody molecule. |
in combination with one another, define the specificity of the antibody for an antigen
|
|
|
7. Which of the following statements about immunoglobulin is true?
A) The ratio of IgA to IgG is higher in the intestinal lumen than in serum. B) Both IgA and IgE can cross the placenta. C) Immunoglobulin heavy chain class switching requires the function of the Recombinase Activating Genes (RAGs). D) The structure of IgA is the same in both saliva and serum. |
****
|
|
|
8. During the rearrangement of immunoglobulin genes
A) an unsuccessful rearrangement on one chromosome may be followed by the successful rearrangement on the other chromosome. B) a successful rearrangement of one chromosome does not prevent the subsequent rearrangement on the other chromosome. C) two nonfunctional rearrangements of the heavy chain gene is usually followed by a switch to a different isotype. D) every nucleotide at recombination points of the heavy chain are encoded in germline sequence the one of genetic elements |
**
|
|
|
9. Immunoglobulin heavy chain class switching from IgM to IgA:
A) occurs frequently during T-independent antibody responses. B) requires the function of Recombination Activating Genes (RAGs). C) involves genetic recombination at the immunoglobulin heavy chain locus. D) is driven by interaction between a B cell and an antigen-presenting cell. E) involves differential splicing of a single large RNA transcript to produce messages encoding either heavy chain. |
**
|
|
|
10. Which of the following are antigen-independent steps in B cell differentiation?
A) Expression of surface IgM. B) Expression of surface IgG. C) Affinity maturation. D) Secretion of large amounts of soluble Ig molecules. E) Both A and C are correct. |
**
|
|
|
11. In a pre-B cell, one would find:
A. cell surface IgM. B. rearrangements between VH, DH and JH segments of Ig genes. C. cytoplasmic mu chains and light chains. D. productively rearranged VL,JL segments of Ig genes. |
**
|
|
|
12. Which of the following statements concerning Ig genes are true?
A. Each light chain isotype is encoded in a separate gene cluster. B. Lambda and the kappa light chain are both present on each Ig C. The constant regions for the heavy chains are encoded on a different chromosome from the variable regions of the heavy chain. D. The complete protein sequence of a VL or VH domain is encoded in a single gene segment. |
**
|
|
|
13. Which of the following statements concerning Ig genes are true? Multiple right answers
A. There are separate exons for EACH V-domain and for EACH of the C-domains. B. Both light chain and heavy chain genes contain J segments. C. Both light chain and heavy chain genes contain D segments. D. Light chains determine antigen specificity |
**
|
|
|
14. Which of these statements is false? Immunoglobulin heavy chain gene transcription:
A. results in a precursor RNA that contains both introns and exons of the entire variable and constant region sequences. B. may result in a single mRNA transcript that contains the VHDJH segment together with both mu and delta constant gene segments. C. is initiated under the control of specific enhancer sequences. D. does not occur in pre-B cells. |
**
|
|
|
15. Short answer: List the 5 antigen-independent and 2 antigen-dependent sources of antibody diversity.
|
**
|
|
|
What encodes the CDR3 region?
|
VDJ and VJ coding joints encode
CDR3 region of VH and V |
|
|
Explain the roles of nonamers and heptamers
|
Nonamers hold gene segments together, heptamers allow
binding of RAG proteins |
|
|
TdT randomly adds nucleotides at the _______ joint
n Greatly increases diversity |
coding
|
|
|
_____and _______ are unique to B and T lymphocytes and essential
for recombination |
RAG and TdT
|
|
|
H and L chains associate to form IgM
|
**
|
|
|
How many unique IgM receptors are produced per day?
|
Approximately 10^7 new B cells,
each with a unique IgM receptor, are produced in the bone marrow every day. |
|
|
When would Repeat attempts at rearrangement
can occur at light chain loci? |
This occurs when the
first rearranged light chain is unable to pair with the rearranged heavy chain |
|
|
What is the downside to gene arrangment?
|
Gene rearrangement is error prone: many cells are lost
|
|
|
How are many cells lost in gene arrangment?
|
errors render the B cells ineffective and they are destroyed ???
|
|
|
Describe the selection of immature B cells
|
Central tolerance is where B cells with antibodies on the surface which recognise “self” antigens are deleted or inactivated. Either through:
- clonal deletion - RAG is reactivated - Anergy |
|
|
Describe clonal deletion
|
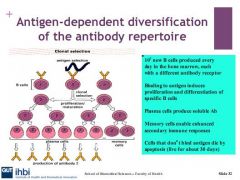
clonal deletion occurs when the b cells have surface igs which have strong affinity or recognise self ags. they are destroyed or inactivated by apoptosis, or inactivated by reapplication of RAG, or undergo Anergy
|
|
|
How many b cells are produced by the bone marrow?
|
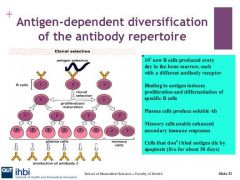
10^7
|
|
|
What stimulates differentiation and proliferation in b cells?
|
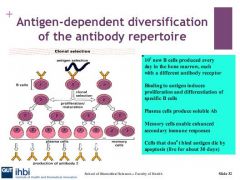
receptor of Ag is internatlised and processed by the b cell and presented to a TH2 (T helper cell) IL2,4,5,6 is expressed which makes B cell differentiate IL4 on the TH2 makes the B cell divide.
|
|
|
Describe the plasma b cell in b cell differenitation and proliferation
|
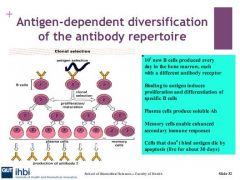
B cells will produce plasma cells which produce immunoglobulins. Plasma cells have no surface Igs and have a short life span
|
|
|
Describe memory b cells
|
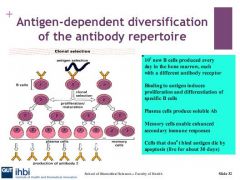
Memory B cells undergo phenotypic changes which make B cells more efficient at recognising Ags, they live longer than b cells and plasma cells - for many years. B cells enable secondary immune responses eg: vaccines
|
|
|
What enable secondary immune reponses?
|
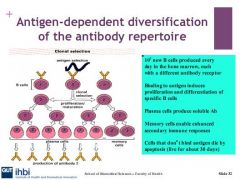
Memory B cells
|
|
|
Which B cells die via apoptosis
|
1. cells which react with self antigens
2. cells which don't bind an antigen within 30 days |
|
|
What are the Three important events during
antigen-dependent B cell development? |

|
|
|
Explain how Membrane-bound and secreted antibodies are derived by alternative RNA processing
|
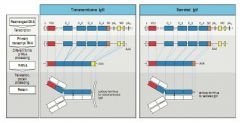
Heavy chain C genes have 2 polyadenylation sites pAs and pAm Polyadenylation at pAm (removal of SC sequence) generates transmembrane IgM
Polyadenylation after pAs generates secreted IgM |
|
|
What encodes membrane bound IgG
|
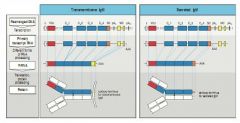
MCD encode transmembrane region of the cytoplasmic tail of the transmembrane igG
|
|
|
What encodes secreted igG?
|
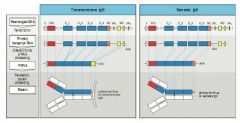
SCD
|
|
|
Which splicing sites will result in differentiation forms?
|
- pAs and pAm
- if splicing occurs between last constant mu exon, and the 5' end of the MC exons, removal of the SC sequence results in the joining of Mc exons and generating a transmembrane Ig. |
|
|
describe Diversification through somatic hypermutation (SHM)
|
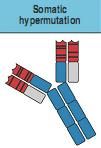
somatic mutation is a form of secondary diversification. it occurs in activated B cells. SM aims to enhance Ab affinity for Ag through point mutations on recombined VDJ region of VH chains in
dividing germinal center B cells |
|
|
Which B cells experience somatic hypermutation
|
dividing germinal centre B cells
|
|
|
Descrive Involves the enzyme activation-induced
cytidine deaminase (AID) |
Results in B cells with both higher and lower
affinity for antigen, process not completely understood |
|
|
Describe the process of AID and other enzymes
|
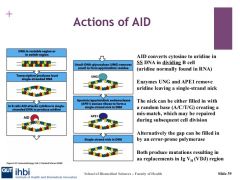
AID binds to the VDJ region and removes the amino group in single stranded DNA, and transforms cytidine to uridine, this introduces single point mutations.
|
|
|
What are the two enzymes with accompany AID?
|
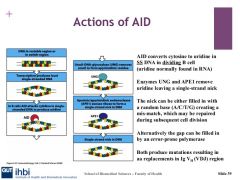
UNG and APE1
|
|
|
What does UNG stand for and perform?
|
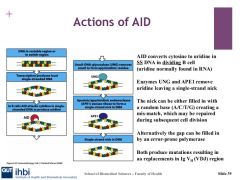
When AID converts cytidine to uridine, it is a foreign element in DNA and a mismatch with Guasonine. DNA repair can be undertaken by base excision repair involving uracil-DNA glycosylase. which makes uracil--> uridine to create a basic site for DNA. This means we have either a random allocation of base opposite abasic site.
|
|
|
What does APE1 stand for? What role does it perform?
|
apurinic/apyramidimic endonuclease1 excises abasic site after UNG and makes a single strand nick which spurs on the reaction of homologous recombination which can results in gene conversion, important for class switchin
|
|
|
Describe the outcomes of somatic hypermutation
|

Overall the fine tuning of immune response which results in high affinity B cells being selected over low affinity B cells (ab) as plasma cells, or memory cells. B cells with low affinity antibody die by apoptosis
|
|
|
What is the rate of mutation for AID?
|
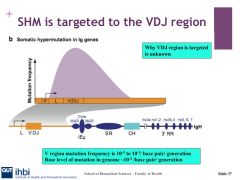
|
|
|
What is the mutation rate for the genome?
|
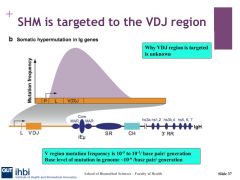
|
|
|
What region does the SHM target?
|
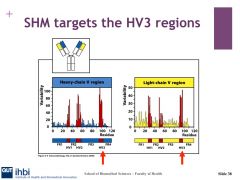
|
|
|
What is class switching recombination?
|
The changing of heavy chain isotyope from constant mu, to another heavy chain isotope during the secondary immune response. While maintaining the VJ and ADJ sequences
|
|
|
How does class switching occur?
|
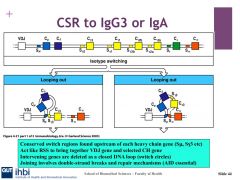
VDJ and CH gene are united through RSS-like activity of the conserved switch regions, which marry VDJ and CH regions via ds breaks and AID splicing out CH genes of non interest to extrachromosomal DNA.
|

