![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
26 Cards in this Set
- Front
- Back
|
Cell Differentiation
|
Cell fate maintained by differential methylation of DNA. All subsequent daughter cells have same amount of methylation site. At fertilization, gDNA is demethylated, and new wave of methylation is initiated → establishes blueprint
|
|
|
X-inactivation (epigenetic)
|
One X-chromo in each somatic cell = inactivated → Barr Body, allows for gene dosage compensation. This is random.
In embryo, both X-chromo are active in females. At implantation, one is randomly inactivated and all daughter cells inherit the methylation pattern → causes mosaicism, 1/2 of cells express paternal alleles and 1/2 express maternal alleles. During gametogenesis, methylation patterns are erased and both X-chromo are reactivated |
|
|
Metabolic Imprinting
|
Epigenetic programming of metabolism during pre-natal/neo-natal periods via in utero exposures (chemical/maternal diet).
Differential methylation causes differential expression of specific genes throughout development and into adult life. This is the only one that can be transmitted to future offspring (transgenerational heritance) |
|
|
Genetic Imprinting
|
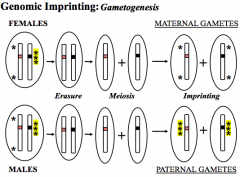
Certain genes expressed in a parent-of-origin-specific manner and can tell which gene came from which parent
CDKN1C = allele from mother IGF2 = allele from father Sperm - paternal imprint on every gene/chromo Oocyte - maternal imprint on every gene/chromo Somatic Cells - imprinting maintained throughout life. Germline cells - erased, reestablished during gametogenesis Mutant allele active on paternal chromo, inherited from mother - no effect Mutant allele active on paternal chromo, inherited from father - effect |
|
|
Beckwith-Wiedemann Syndrome
|

Disorder of growth - characterized by large size for gestation age, large tounge, abd. wall defects, predisp to embryonic tumors
Caused by 2 active copies of IGF-2 gene and/or no active copy of CDKN1C CDKN1C = allele expressed only from maternal inherited chromosome IGF2 = allele expressed only from paternal inherited chromosome Loss of Methylation CDKN1C mutation Uniparental Disomy |
|
|
Prader-Willi Syndrome and Angelman Syndrome
|
PWS = short stature, hypotonia, small hands/feet, obesity, mild-moderate mental retardation, hypogonadism
AS = severe mental retardation, seizures, ataxic gait PWS = inherit deletion from father, 5 genes, active only on paternally derived chromosome, neither copy of 15q has paternal imprinting AS = inherit deletion from mother, UBE3A, active only on maternally derived chromosome, neither copy of 15q has maternal impirinting Uniparental Disomy 2 normal copies of maternal chromo 15 leads to PWS (normal PWS genes, but silent on both) 2 normal copies of paternal chromo 15 leads to AS (normal AS genes, but silent on both) |
|
|
PWS vs. AS
|
Inherit deletion from father, failure to demethylate = PWS
Inherit deletion from mother, failure to methylate = AS |
|
|
Anticipation
|
Occurs in some dominant disorders
⬆ severity in successive generations (correlated w/# of repeats) and earlier onset Congenital form seen only in infants of affected mothers Caused by repeat expansion Repeat is unstable and varies each generation Expansion occurs during gametogenesis in females |
|
|
Triplet Repeat Expansion
|
Trinucleotide repeats are repeated a variable but low number of times.
Each allele varies w/in normal range of each generation If repeat expands beyond normal range it becomes unstable and expands in subsequent generations (premutation) Anticipation also occurs |
|
|
Myotrophic Dystrophy
|
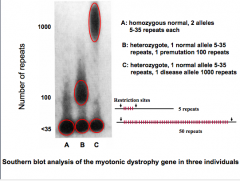
Most common muscular dystrophy in adults
Repeat expansion in protein kinase RNA ➔ decreases RNA stability Autosomal dominant Expansion occurs during gametogenesis in females - will only see congenital form in infants of affected mothers |
|
|
Huntington Disease
|
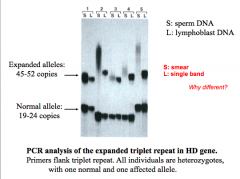
Repeat expansion in coding region of Huntington protein ➔ gain of fxn mutation
When expansion is past premutation, it is transmitted autosomal dominant Full mutation is 100% penetrant Triplet repeats occur in frame w/in coding region = ⬆ number of glutamines Greater expansion when transmitting parent is male |
|
|
Fragile X Mental Retardation
|
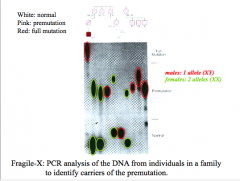
Most common form of mental retardation
FMR1 RNA binding protein ➔ inhibits transcription due to hypermethylation X-linked dominant (milder/variable expression in females than males) 80% penetrant in males 20% penetrant in females Caused by compensation of X in females Unaffected sisters of affected siblings = high risk of affected child Unaffected brothers = no risk of affected child Unaffected daughters of NTM = high risk of affected child Full mutation = 100% penetrant in males, 50% penetrant in females Expansion occurs exclusively through mother Females are at risk of premature menopause Males are at risk of neurodegenerative disorder |
|
|
Mitochondrial mutations
|
Deficient ox. phos. in heart, skeletal, and CNS. Myopathies (muscle), neuropathies (neurons), encephylopathies (brain)
Higher rate of mutation due to reactive oxidation species and DNA pol in mito is less accurate and not corrected efficiently Mitochondria also depend on nuclear DNA for essential proteins (Mitochondria loss of fxn can occur from nuclear DNA mutation, will not be strictly maternal inheritance) Maternal inheritance - all mitochondria come from the oocyte, affected males do not transmit to offspring |
|
|
Mitochondrial inheritance w/o heteroplasmy (LHON)
|
Only females transmit mitochondria
Affected female = all children are affected Affected male = no children are affected Leber's Hereditary Optic Neuropathy Delayed age of onset - accumulation of mtDNA mutations (1 of 3 missense mutations) |
|
|
Factors that Affect Mitochondrial Fxn
|
Inherited capacity for ox phos - both nucelar and mito genes
Tissue specific requirements for ox phos Age - capacity for ox phos decreases w/ age (accumulation of mtDNA mutations) Accumulation of somatic mtDNA mutations and degree of heteroplasmy |
|
|
Mitochondrial inheritance w/ Heteroplasmy (MERRF/MELAS)
|
Mutant tRNA = incorrect aa inserted into all mito proteins w/ this codon
More mutant tRNA:normal tRNA = more severe disorder MERRF (myoclonic epilepsy w/ragged-red fiber) Myoclonus, epilepsy, ataxia, ragged-red fiber in muscle biopsy Single base changes in tRNA = changes codon specificity Maternal transmission Highly variable expression due to heteroplasmy |
|
|
Heteroplasmy
|
Mosaicism caused by variable number of mutant mitochondrial/cell
Causes variable expression of disease/tissue specific differences from person to person Variation arises from Genetic drift (chance variation) - just the nature of miDNA replication Selective advantage - deletion allows for faster replication |
|
|
Replicative Segregation
|
Mito replicate by division w/in somatic cell
Prior to division, mtDNA are replicated a variable number of times Each mito inherits a variable number of mt chromosomes |
|
|
Mitotic Segregation
|
Each cell has a population of different mito, that are randomly distributed throughout cell
When daughter cells form by cytokinesis - they randomly inherit a variable number of mito which replicate after cell division |
|
|
Multifactorial/Polygenic
|
Genetic, but needs a trigger or is influenced by envrionment
|
|
|
Diabetes
|
Susceptibility is genetic but envrion. trigger is required
Twin studies show that susceptibility to obesity/diabetes is genetic Pima Indian studies show that envron. trigger is needed |
|
|
Heritability
|

Concordance Rates in Twins = % of time both inherit a trait
If 100% genetic: MZ twins = 100%, DZ twins 50% If 0% genetic: MZ twins = DZ twins = sibling concordance Heritability - portion of total variance caused by genes If not 100%, then there are strong environmental triggers |
|
|
Recurrence and Transmission patterns
|
Risk can't be calc. based on probability of shared genes
Recurrence risk determined from empirical data (observation of pop). This will change from pop to pop and from family to family (differences in genetic load) Recurrence risk decreases rapidly for more distantly related relatives increases if more than one family member affected (higher genetic load) more severe expression = higher risk (higher genetic load) increased if proband is of less commonly affected sex (higher genetic load) If trait is genetic = risk is proportional to coefficient of relationship If trait is multifactorial = risk decreases more rapidly than predicted from coefficient of relationship |
|
|
Quantitative Trait
|
Phenotype distributes across a gaussian curve across a population, but sp. values for trait cluster in families
More genes/envrionmental factors involved, the better the curve |
|
|
Threshold of Liability
|
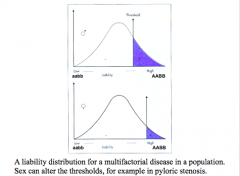
Disease does not have normal distribution but is a result of continuous underlying liability.
If threshold is exceeded, then disease results. Even if you have an environmental trigger, if it's not above the threshold, no disease will occur Less alleles to be affected as a male More alleles to be affected as a female Who would have greater risk, siblings of affected female or an affected male? Affected female - parents have a higher genetic load |
|
|
Genetic load
|
Number of disease susceptibility alleles someone carries
|

