![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
52 Cards in this Set
- Front
- Back
|
Epigenetics |
A change in either a chromosome structure or a gene structure |
|
|
Effects/results of epigenetics |
-Affects the phenotype -not a permanent change -not a mutation -not a change in DNA sequence |
|
|
Two major types of epigenetics |
Dosage compensation and Genomic imprinting |
|
|
Dosage Compensation |
Embryogenesis (occurs after fertilization) (x chromosome inactivation) |
|
|
Genomic Imprinting |
Gametogenesis (occurs before fertilization) |
|
|
Lyon Hypothesis |
suggesting that doseage compensation in mammals is by inactivation of all but one X chromosome in cells with more than one X chromosome |
|
|
How are chromosomes counted? |
By the x chromosome inactivation center (XiC). The first xic is counted and remain 'on', when the second is counted it is turned 'off'. |
|
|
The XiC contains two genes (______ and _______) and one control region (________). |
Genes: Xist and Tsix Control Region: Xce |
|
|
How do Xist and Tsix work together? |
Both are nonstructural genes (they don't produce proteins). Xist produces a functional RNA molecule that recruits proteins that make a barr body while Tsix produces RNA that counteracts Xist. When the two RNA strands combine they form a dsRNA that is recognized as a viral invasion by the body and is then destroyed, so the barr body producing Xist is destroyed in the process. |
|
|
Xce Control Region |
-A DNA sequence that regulates the Tsix gene -Protein binding site |
|
|
Genomic Imprinting |
Epigenetic phenomenon by which certain genes are expressed in a parent-of-origin-specific manner. If the allele inherited from the father is imprinted, it is thereby silenced, and only the allele from the mother is expressed. |
|
|
Your genotype does not determine your ______. |
Phenotype |
|
|
Methylation of "C" bases does what? |
Turns genes off |
|
|
With DNA replication in bacteria, the template strand is read in the ________ direction and the new strand is synthesized __________. |
3' to 5' 5' to 3' |
|
|
Structural vs Non-Structural genes |
Structural genes produce mRNA that will code for proteins, non-structural genes encode RNA's that are never translated |
|
|
In conservative replication, the original strand _____ |
stays together |
|
|
In semi-conservative replication, the original strand _____ |
splits |
|
|
In dispersive replication, the original strand _____ |
breaks apart |
|
|
The Meselson-Stahl experiment used this bacteria which replicates every _____ minutes |
E. Coli; 20 minutes
|
|
|
What was used in the Meselson-Stahl experiment to mark the original and synthesized strands? What solution was it placed in? |
Nitrogen 15 (heavy) = original DNA strand Nitrogen 14 (light) = newly synthesized strand Dense solution of cesium chloride (CsCl) |
|
|
Results of Meselson-Stahl experiment |
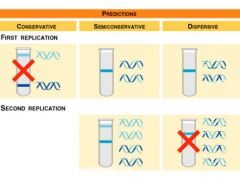
|
|
|
What are the four DNA replication rules? |
1) newly synthesized strand made 5' to 3' 2) template strand is read 3' to 5' 3) leading strand made in same direction as replication fork 4) lagging strand made in opposite direction of the replication fork |
|
|
Initiation DNA sequence for DNA Replication in E Coli |
OriC |
|
|
Initiation Proteins for DNA Replication in E Coli |
DnaA DnaB (DNA Helicase) ssBP's DNA adenine methyl-transferase (DAM) |
|
|
Elongation Proteins for DNA Replication in E Coli |
DNA Helicase SSBPs DNA gyrase (topoisomerase II) DNA primase DNA polymerase III holoenzyme DNA polymerase I DNA ligase |
|
|
Termination Proteins for DNA Replication in E Coli |
Tus DNA Ligase DNA gyrase |
|
|
Termination Sequences for DNA Replication in E Coli |
Termination (ter) sequences |
|
|
What are three reasons that DNA replication in Eukaryotes is more complex than in bacteria |
1) larger genomes 2) linear chromosomes 3) multiple origins |
|
|
Initiation Sequences for DNA Replication in Eukaryotes |
ARS Element |
|
|
Initiation Proteins for DNA Replication in Eukaryotes |
Origin Replication Complex (OrC) Cdc 6/ Cdt 1 MCM helicase |
|
|
Elongation Proteins for DNA Replication in Eukaryotes |
MCM Helicase Replication protein A (RPA) DNA Primase/ DNA polymerase alpha complex Topisomerase II DNA polymerase epsilon DNA polymerase delta Fen1 DNA ligase I PCNA |
|
|
Telomeres |
form t-loop structures due to unusual hydrogen bonding between guanine bases (G quartet) |
|
|
Telomerase |
Solves the shortening of linear chromosomes in replication by adding a 5'-TTAGGG-3' sequence to the 3' ends of template strands. Has a built in RNA template which is used to synthesize DNA and is considered a DNA polymerase. Without telomerase, cells would die after roughly 50 divisions due to too great a loss of genetic information. |
|
|
Polymerase Chain Reaction (PCR) |
DNA replication outside a living organism (test tube). Used in forensics, analysis of genes, evolutionary studies, etc. |
|
|
PCR setup: |
Place into a test tube: -template DNA -two types of DNA primers (customized to the strand) -taq DNA polymerase -dNTP's -buffer (maintains reaction conditions) |
|
|
PCR thermocycler stages: |
1) Denaturing (95 C for 1 min) 2) Primer Annealing (55 C for 1 min) 3) DNA Synthesis (72 C for 2-3 mins) |
|
|
How many copies of gene X are present at a given time during PCR? |
a(2^n) a: number of template strands n: number of cycles |
|
|
How do we know that PCR worked? |
Agarose Gel electropheneisis -allows you to visualize DNA -allows you to separate DNA molecules by size |
|
|
Agarose Gel Electrophenesis procedure |
1) Dissolve agarose in a buffer (use microwave) 2) Allow to cool 3) Add ethidium bromide dye to gel before it solidifies 4) Pour gel into casting tray 5) Add comb to create wells 6) Allow to solidify 7) Load samples 8) Apply electric field (DNA migrates to + electrode) 9) Visualize Gel |
|
|
What travels further in Agarose gel electropheneisis? Large or small template strands? |
The smaller the strand, the further it travels. |
|
|
Transcription in Bacteria: Initiation overview |
Involves binding of a transcription factor to the promoter. Recruitment of RNA polymerase |
|
|
Transcription in Bacteria: Elongation overview |
RNA polymerase separates the strands (open complex) Synthesis of an RNA transcript |
|
|
Transcription in Bacteria: Termination overview |
Release of the transcript Release of RNA polymerase |
|
|
In transcription, the 5' to 3' strand is the _______ or ______ strand, while the 3' to 5' is the ______ or ______ strand. |
Coding or sense Template or antisense |
|
|
The ______ strand has the same sequence as the mRNA (but has 'T' instead of 'U') |
coding/sense |
|
|
Initiation Sequences for DNA Transcription in E Coli
|
1) Regulatory DNA sequences 2) Promoter |
|
|
Initiation Proteins for DNA Transcription in E Coli |
1) Sigma factor 2) RNA Polymerase |
|
|
Elongation for DNA Transcription in E Coli |
A mRNA is synthesized by RNA polymerase |
|
|
What are the two processes of Termination of Transcription in E Coli |
Rho-dependent termination Rho-independent termination |
|
|
What is a stem-loop? |
Two groups of GCGC sequences on a single mRNA strand bond together to form a stem with the base pairs between the two groups forming a loop. This structure causes the RNA polymerase to stall |
|
|
Rho-Dependent Termination |
1) RNA polymerase transcribes gene X 2) Transcription generates rut sequence in mRNA 3) Rho protein binds to rut in the mRNA - rho protein moves along mRNA 5' to 3' - rho protein is a helicase that separates RNA from DNA 4) The stem-loop in the mRNA causes RNA pol to stall/slow down 5) Rho protein catches up to RNA polymerase 6) Rho protein separates mRNA from template DNA strand 7) RNA polymerase falls off, transcription terminates |
|
|
Rho-Independent Termination
|
1) RNA polymerase transcribes gene Y 2) RNA pol stalls at the stem-loop -stalling may be assisted by NusA 3) When RNA pol stalls, it is transcribing a U-rich sequence in the mRNA 4) U-A base pairs spontaneously break 5) RNA polymerase and mRNA are released 6) Transcription terminates |

