![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
54 Cards in this Set
- Front
- Back
|
Process of protein synthesis
|
a. mRNA enters cytoplasm, associates with ribosome
b. mRNA aligns itself on ribosome so first codon is read 1st c. tRNA w/ complementary amino acid to 1st codon binds to 1st codon (tRNA with AA for 2nd binds to 2nd codon, etc.) d. mRNA codons continue to be read toward carboxyl terminus e. protein is released from mRNA and ribosomal particle disassembles |
|
|
Releasing factors
|
a. Help terminate protein synthesis
|
|
|
Signal peptide
|
a. Amino acid sequence that signals where the protein will be targeted
b. 10-20 amino acids at amino terminal |
|
|
Structure and function of tRNA
|
a. Folded into a cloverleaf structure
b. Accomplished by H-bonding between complementary bases |
|
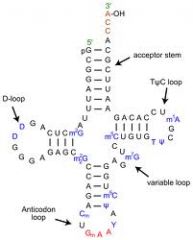
tRNA regions
|
a. 3’ end of molecule→ terminates in CCA→ amino acid binding site
b. TψC loop→ tRNA binds to ribosome c. D loop→ contains a high percentage of dihydrouridine residues (recognized by tRNA synthetase) d. Anticodon loop→ loop contains the anticodon which is complementary to the codon of the messenger RNA a. 3’ end of molecule→ terminates in CCA→ amino acid binding site b. TψC loop→ tRNA binds to ribosome c. D loop→ contains a high percentage of dihydrouridine residues (recognized by tRNA synthetase) d. Anticodon loop→ loop contains the anticodon which is complementary to the codon of the messenger RNA |
|
|
Possible functions of rare or minor bases in tRNA
|
a. Disallow base pairing at certain points in order to form cloverleaf structure
b. Disallow unwanted base pairing with the mRNA c. Protect the tRNA molecules from the action of ribonucleases |
|
|
Isoacceptor tRNA
|
i. Greater than 20 types of tRNA because of redundancy of amino acid code
ii. Recognize several different codons, but all recognize a single amino acid |
|
|
Base pairing
|
i. Anti-parallel
ii. tRNA→ 3’-5’ iii. mRNA→ 5’-3’ |
|
|
Wobble hypothesis
|
i. Third nucleotide of the codon is degenerate
ii. U in anticodon can recognize A or G iii. G in anticodon recognizes C or U iv. I in anticodon recognizes U, C, or A |
|
|
Inosine
|
i. Formed by adenosine deaminase after tRNA is formed on certain tRNAs
|
|
|
Tertiary structure of tRNA molecules
|
i. L-shaped
|
|
|
Activation
|
a. Involves coupling of an amino acid to its corresponding tRNA
b. Catalyzed by amino acyl tRNA synthetases c. Ester bond between amino acid and tRNA molecules is a high-energy bond→ AA bound to 2’ or 3’ -OH group of ribose of terminal residue of tRNA d. Activation requires equivalent of 2 ATPs |
|
|
Aminoacyl-tRNA synthetase
|
i. Specific to tRNA and amino acid
ii. 20 tRNA synthetases iii. Recognize multiple points on tRNA structure |
|
|
Error correction
|
1. Occurs while tRNA is bound to the synthetase
2. Editing mechanism where correct tRNA is recognized and corrected by the synthetase itself 3. One protein per thousand will contain incorrect AA due to misacylation of tRNA |
|
|
Role of amino acid in recognition of anticodon for amino acid
|
a. NONE
b. If an amino acid attached to tRNA is chemically altered, the altered form will be incorporated into the protein |
|
|
Ribosomes in initiation
|
i. Both prokaryotic and eukaryotic ribosomes consist of 2 subunits
ii. Each subunit is composed of proteins and rRNA |
|
|
Initiation process
|
i. Initiation factors dissociates the 70S (80S in eukaryotes) ribosomes into 50S and 30S (40S and 60S)
ii. tRNAf met (formyl methionine) is the initiation codon for prokaryotes→ binds to IF2-GTP (required for binding to 30S subunit) iii. 50S reassembles with 30S and tRNAf met introduces the first amino acid→ binds to initiation codon at P site, IF2 is lost from complex with GTP hydrolysis |
|
|
IF 1, 2, 3
|
1. IF1 and 3 bind to free 30S subunit
2. mRNA then also binds to 30S ribosome subunit 3. IF2 (GTP) binds to tRNAf met which binds AUG at the P site 4. IF2 is lost as GTP is hydrolyzed and 70S ribosome is formed and IF1 and 3 are lost |
|
|
e1F-4E
|
mRNA cap binding factor which is essential for lining up the start codon on the ribosome
|
|
|
A&P (E sites)
|
i. P=peptide site
ii. A=acceptor site iii. E=exit site for tRNA ejection |
|
|
Start codon recognition
|
i. AUG in both pro and eukaryotes
ii. Protein synthesis begins by positioning AUG codon in P site with tRNAf met or tRNA met bound to it iii. mRNA is translated starting at AUG at the 5’ end of mRNA |
|
|
Shine-Delgarno sequence
|
1. AGGAG
2. Found between 5’ end of mRNA and initiation codon 3. Permits binding and proper orientation of mRNA with ribosome |
|
|
Eukaryotic start codon
|
i. AUG nearest 5’ cap
ii. 1 per mRNA molecule iii. Only one protein can be made per eukaryotic mRNA molecule→ monocystronic |
|
|
Kozac consensus sequence
|
i. Determines relative efficiency of eukaryotic message translation
ii. Messages in close proximity to Kozac sequence will translate efficiently |
|
|
Elongation binding
|
i. Aminoacyl tRNA possessing the anticodon to the codon in A site forms complex with EF-Tu and GTP
ii. Complex binds to ribosome such that aminoacyl tRNA is positioned at A site iii. GTP of EFTu-GTP complex is hydrolyzed and EFTu is released |
|
|
Peptide bond formation
|
i. Formed by nucleophilic attack of alpha-amino group of AA in A site on ester bond between AA and tRNA in P site
ii. Catalyzed by ribosomal RNA iii. Energy fro synthesis from activated amino acid attached to tRNA |
|
|
Translocation
|
i. Movement of ribosome one codon on mRNA with release of empty tRNA present in P site and transfer of petide-bearing tRNA from a to P site
ii. Requires elongation factor EFG and GTP hydrolysis iii. A site becomes vacant and the next amino acid charged tRNA enters and binds iv. On average, 10 EF-Tu-GTP amino acyl tRNAs are tried in the A site before the correct one matches |
|
|
Termination
|
i. Termination codon reaches A site (UAA, UAG, UGA)
ii. Releasing factors RF1, RF2, and RF3 causes release of polypeptide chain from ribosome iii. Releasing factors cause peptidyl transferase activity to switch to a hydrolase activity to release the completed polypeptide |
|
|
Polycistronic mRNA
|
i. In prokaryotes, if AUG is not too far from terminal codon, 70S will not dissociate but will form a new initiation complex which will lead to translation of second protein
ii. One mRNA can code for several proteins and each one might be translated |
|
|
Polyribosomes
|
a. As ribosome translates the mRNA, the 5’ end becomes exposed
b. Second initiation complex can form c. Distance between ribosomes is about 100 nucleotides |
|
|
Activation energetics
|
i. Synthesis of AA tRNA uses 2 ATP equivalents for each AA introduced to the protein
ii. Energy of tRNA-AA bond drives peptide bond formation |
|
|
Energetics of elongation and translocation
|
i. 2 GTP/AA
ii. 1 GTP per residue at A site iii. 1 GTP per translocation iv. Energy used to change conformation of translation machine and not for forming new covalent bonds |
|
|
Prokaryotes v. eukaryotes-- initiation
|
i. Initiation amino acid in eukaryotes is methionine
ii. Regulation occurs by phosphorylation of eIF-2→ translation inhibition |
|
|
Pro v. eu-- factors
|
i. Eukaryotic cells require more initiation and elongation factors than prokaryotic cells
|
|
|
Pro v. eu-- 5'-cap
|
i. Binding of mRNA to 40s subunit requires 5’-cap
ii. Proper placement of 5’-cap requires cap binding protein CBP (eIF-4F) |
|
|
Pro v. eu-- cistronic
|
i. Eukaryotic mRNA is monocystronic
ii. Prokaryote mRNA is polycystronic |
|
|
Pro v. euk--- ribosomes
|
i. Eukaryotic cells have both free and bound ribosomes
ii. Prokaryotes have only free ribosomes |
|
|
Pro v. euk-- signal peptide
|
i. Signal for a ribosome to attach to membrane of ER is in synthesized protein
ii. Found in first 15-30 AA residues |
|
|
Protein trafficking
|
i. Many proteins of eukaryotic cells must be transported across the ER or integrated into the ER
ii. Proteins destined for secretion follow this pathway |
|
|
SRP
|
i. Signal recognition particle binds to signal peptide as it exits the ribosomal complex
ii. SRP blocks translation until ribosome has docked w/ ER membrane iii. SRP contains 6 proteins and an RNA molecule as central component |
|
|
SRP receptor
|
i. Found only in ER
ii. Binds SRP when it is part of ribosomal complex iii. GTP binds to SRP and SRP receptor→ protein targeting iv. Forms a gated channel which opens upon ribosome docking |
|
|
Ribphorin
|
i. Ribosomal receptor binds the ribosome
|
|
|
Translocation
|
i. Signal peptide enters ER lumen
ii. Protein elongation continues iii. Signal peptide is removed by signal peptidase iv. ATP required to transport protein through the membrane |
|
|
Secreted protein
|
i. Proteins destined for secretion enter ER lumen and are transported to Golgi for modification and glycosylation
|
|
|
Membrane proteins
|
i. Contain stop-transfer sequences→ signal a stop to translocation of growing peptide chain into ER lumen
|
|
|
Protein synthesis in mitochondria
|
a. Mitochondria contain mtDNA
b. Resembles prokaryotic DNA c. mtDNA codes for nine mitochondrial polypeptides |
|
|
Transport of protein into mitochondria
|
i. Most coded for by nuclear DNA
ii. N-terminal of proteins targeted for mitochondria are post-translationally imported into mitochondria |
|
|
Cycloheximide
|
1. Inhibits peptide bond formation in eukaryotic cells
|
|
|
Diptheria toxin
|
1. Catalyzes the reaction of NAD+ with a eukaryotic elongation factor, resulting in inhibition of translocation
|
|
|
Chloramphenicol
|
1. Inhibits peptide bond formation
2. Can inhibit mitochondrial protein synthesis at high levels pro/euk |
|
|
Puromycin
|
1. Cause premature termination by acting as an aminoacyl-tRNA analogue
2. The AA chain in the P site is transferred to puromycin rather than to the A site pro/euk |
|
|
Tetracycline
|
1. Inhibits the binding of aminoacyl tRNAs to the A site on the ribosome→ inhibits elongation
eukaryotic |
|
|
Streptomycin
|
1. Binds to 30s subunit and causes dissociation of the translation process
prokaryotic |
|
|
Erythromycin
|
1. Binds to the 50s subunit and inhibits translocation
prokaryotic |

