![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
23 Cards in this Set
- Front
- Back
|
how such information stored as nucleic acids can be decoded into protien?
|
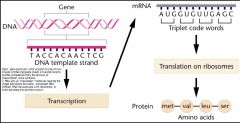
couple question address in chpter about gene expression,
1. how is genetic information encoded? 2. how does the transter from DNA to RNA occur, thus defing the process of transciption? -code are writen in units of three letter-ribonucleotides present in mRNA that reflects the stored information in gene -each triplet code word directs the incorporation of specific amino acid into a protien as it is synthesized. |
|
|
What are the characteristics of genetic codes exhibits?
|
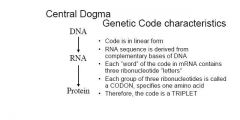
1. genetic code is written in linear form , using the ribonucleotide bases that compose mRNA molec. as 'letters'. the ribonucleotide sequence is derived from the complementary nucleotide bases in DNA
2. Each "word" within the mRNA contains three ribonucleotides letters. each group of three ribonucleotides, called codon, specifies one amino acid 3.the code is unambiguous, meaning that each codon specifies only a single amino acid. 4. the code is degenerate, meaning that given amino acid can be specified by more than one codon. this the case for 18 of the 20 amino acids 5. the code contains "start and 'stop' signals, certain codons that are necessary to initiate and to terminate translation 6. no internal puctuation'commas' is used in the code, thusthe code is said to be commaless. once translation of mRNA begins, the codons are read one after the other with no breaks between them. 7. the code is nonoverlapping. after translation commences, any single ribonucleotides at a specific location within the mRNA is part of only one codon 8. the code is nearly universal. with only minor exceptions a single coding dictionary is used by almost all viruses, prokaryotes, archea, and eukaryotes. |
|
|
What are the Genetic Code characteristics?
|
• Code is in linear form
• RNA sequence is derived from complementary bases of DNA • Each “word” of the code in mRNA contains three ribonucleotide “letters” • Each group of three ribonucleotides is called a CODON, specifies one amino acid • Therefore, the code is a TRIPLET |
|
|
What are the Genetic Code characteristics? cont'
|
Genetic Code characteristics
• The code is unambiguous, each triplet specifies a single amino acid • The code is degenerate, more than one triplet can code for the same amino acid • The code contains “start” and “stop” signals • Translation of mRNA is continuous • The code is nonoverlapping • The code is “nearly” universal |
|
|
How was the code
determined? |
• How does four nucleotides encode the 20 amino acids?
• Evidence supported a nonoverlapping code • 1961: Jacob and Monod postulated the mRNA • mRNA was discovered, the code that is translated is in mRNA |
|
|
What was the code?
|
• Theoretical argument of a triplet
• 3 letters represents the minimum to encode nformation to specify 20 amino acids • 43 =64 different possible combinations • 42= 16 not enough, 44=256 too much |
|
|
First evidence of triplet nature of code was discover by what experiment?
|
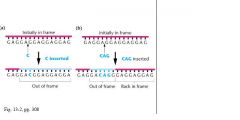
• Francis Crick involved in this as well
• Insertion & Deletion mutations in T4 bacteriophage • Used intercalating agents, get into the stacked bases of DNA causing insertions or deletions upon replication |
|
|
explain the process of first evidence of triplet nature of code?
|

• This insertion or deletion causes the reading frame to shift: frameshift mutation
• Treat the mutants again with intercalating agents could result in reversal of the mutant phenotype • One + and one - : normal phenotype • Also +++ and --- : normal phenotype: supports triplet nature of the code |
|
|
What if Overlapping triplet
code? |
• Consider a sequence: GTACA
• If the triplet code were overlapping, then GTA, TAC, and ACA are possible reading frame codons of this sequence • If overlapping, it restricted the amino acids that would be adjacent to the amino acid encoded by the central triplet • If true, then sequence of tripeptides would be limited, but that was not the case when peptide sequences were examined |
|
|
what was the Other arguments against an overlapping code?
|
• Point mutations would affect more than one amino acid, but this was not observed
• Crick argued for an adaptor molecule, not consistent with an overlapping code • BOTTOM LINE: Evidence did NOT support an overlapping code • CONCLUSION: The code is NONOVERLAPPING |
|
|
how was decide to deciphering the “Code”? and
|

• 1961: Nirenberg and Matthaei characterized the first specific coding
sequences • In vitro system (cell-free) that could synthesize protein • Enzyme (polynucleotide phosphorylase) that produced synthetic mRNAs, which serve as template for polypeptide synthesis in cell free system |
|
|
how was decide to deciphering the “Code”? and methods
|
• Polynucleotide phosphorylase can be
made to synthesize RNA in vitro • The formation of this RNA is random, based on the concentration of the four ribonucleoside diphosphates added to the in vitro system Method • Polynucleotide phosphorylase can be made to synthesize RNA in vitro • The formation of this RNA is random, based on the concentration of the four ribonucleoside diphosphates added to the in vitro system • Therefore; the probability of the insertion of a particular ribonucleotide is proportional to its availability in the system PROVIDES A MEANS TO DECIPHER THE CODE |
|
|
Simplest Experiments done to bring about homopolymers code in deciphering the in running free cell snthesis?
|
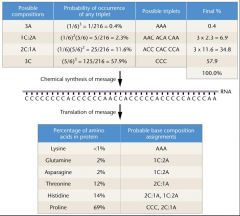
• Homopolymers
• Synthesize RNA homopolymers consisting of only one of the ribonucleotide • Eg., UUUUUUU…, AAAAAAA….., CCCCCC….., GGGGGGGGG……. • Radioactively label each of the 20 amino acids • Determine what amino acids these homopolymers specify, see Table 12-1 |
|
|
what were Homopolymer results?
|

• UUU: Phenylalanine
• AAA: Lysine • CCC: Proline • GGG: no result in these early experiments |
|
|
what did the mixing of the copolymer Heteropolymers achieved?
|
• Next moved to heteropolymers with
known concentrations (proportions) of each ribonucleotide • Could predict the frequency of triplets based on proportions • Match the proportion of amino acids incorporated with the proportion of potential triplets Heteropolymer example • Produce RNA with 1A:5C ratio • Insertion of A or C at any particular position is determined by ratio of A:C • Eg., frequecy of AAA = 1/6 * 1/6 * 1/6 = (1/6)3 = 0.4% |
|
|
what did the mixing of the copolymer Heteropolymers achieved?
|
Heteropolymer example
• Produce RNA with 1A:5C ratio • Insertion of A or C at any particular position is determined by ratio of A:C • Eg., frequecy of AAA = 1/6 * 1/6 * 1/6 = (1/6)3 = 0.4% • Eg., frequency of AAC, ACA, CAA = (1/6)2 * (5/6)= 2.3% each Heteropolymer example • Next: examine the percentages of incorporation of any given amino acid Heteropolymer example • Next: examine the percentages of incorporation of any given amino acid • Propose what triplet might encode that amino acid • Eg., proline seen 69% of the time: = CCC (57.9%) + 2C1A(11.6%), determines the composition of the triplet coding for an amino acid, not the specific sequence of the triplet yet • Next: examine the percentages of incorporation of any given amino acid • Propose what triplet might encode that amino acid • Eg., proline seen 69% of the time: = CCC (57.9%) + 2C1A(11.6%), determines the composition of the triplet coding for an amino acid, not the specific sequence of the triplet yet • Conduct many experiments of this type to work out the code! |
|
|
what did Triplet Binding Assays determine, inaddition the discover works?
|
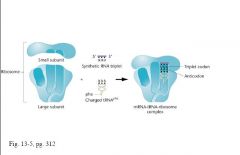
• 1964: Nirenberg and Leder
• Led to specific assignments of triplets • Single triplets could be bound by ribosomes • Leads to binding of anticodon • Experimental technique eventually led to determination of specific triplets coding for 50 of 64 codons • Established degeneracy of the code: >1 codon for 18 of 20 amino acids |
|
|
what are the Repeating copolymers?
|

pictuture
• Di, tri, and tetra repeats • Confirmed codons already established, filled in the remaining gaps • Also established termination signals, code for NO amino acid incorporation |
|
|
Final codon table
|
• 61 triplet codons that specify amino
acids • 3 termination codons, stop amino acid incorporation |
|
|
Degeneracy of Code
|
• Code is degenerate: almost all amino
acids encoded by 2, 3, 4 codons • Three (serine, arginine, leucine) encoded by 6 codons • Only 2 (tryptophan and methionine) encoded by a single codon • 3 termination codes |
|
|
Wobble hypothesis
|
• Pattern of degeneracy
• Third letter of the code is usually the one that is different: 3rd position free to “wobble” • Pattern of degeneracy • Third letter of the code is usually the one that is different: 3rd position free to “wobble” • First 2 positions are most critical • Allows 1 anticodon of tRNA to pair with >1 codon in mRNA |
|
|
The genetic code is “nearly”
universal |
• In some organisms or in mitochondrial
genomes, the codon usage is different • Eg. “normal” UGA is a termination codon, in human & yeast mitochondrial genomes codes for insertion of tryptophan • Other exceptions: Mycoplasma, Paramecium, Tetrahymena, & a few other organisms |
|
|
Overlapping Genes
|
• Codons are still NOT overlapping
• Start initiation of transcription at different locations • Result is different reading frames produce more than one polypeptide • Seen in viruses, eg. φX174 |

