![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
259 Cards in this Set
- Front
- Back
- 3rd side (hint)
|
The functional unit of genetic information
|
gene
|
|
|
|
In all cells genes are composed of
|
deoxyribonucleic acid (DNA)
|
|
|
|
DNA is transferred to
|
RNA
|
|
|
|
The central dogma of genetic information flow
|
DNA to RNA to protein
|
|
|
|
The processes that make the information flow:
|
1.) Replication
2.) Transcription 3.) Translation |
|
|
|
four nucleic acid bases
|
Adenine, Thymine, Guanine, Cytosine
|
|
|
|
The backbone of DNA is a repeated pattern of
|
Phosphate, pentose deoxyribose sugar, base.
|
|
|
|
DNA is complementary, meaning?
|
the two opposite bases on different strands are complimentary
|
|
|
|
The two strands of DNA are Antiparallel, meaning?
|
One strand being read 5' to 3' is the same as the complimentary strand read 3' to 5'
|
|
|
|
The two strands form what kind of structure:
|
Double helix.
|
|
|
|
Major groove and minor groove?
|
Major grooves lead to more interaction. More space between molecules for interaction
|
|
|
|
Long DNA are quite flexible and stem loop strucures form because of protein interaction with these pattersns
|
inverted repeats
|
|
|
|
Such secondary stricture as stem loop are critical to the function of
|
tRNA
|
|
|
|
DNA bases are bonded together by
|
Hydrogen bonding (individually weak but are stabilizing)
|
|
|
|
If the heat is raised what happens to DNA
|
the heat breaks the hydrogen bonds and the strands seperate
|
|
|
|
If the heat is then removed the strands come together known as
|
annealing.
|
|
|
|
Negative supercoiling
|
coiling in the opposite direction of the right handed double helix.
|
|
|
|
In eukaryotes a lot of protein bound to DNA known as __________ to form structures known as _______.
|
histones; nucleosomes
|
|
|
|
Loose genetic material is knwon as __________ vs. when it is tightly wound known as?
|
chromatin; chromosome
|
|
|
|
the enzyme which introduces negative supercoils is
|
DNA Gyrase
|
|
|
|
DNA gyrase is a type of enzyme known as a
|
Topoisomerase
|
|
|
|
The enzyme which removes supercoils
|
Topoisomeras 1
|
|
|
|
Supercoiling besides replication also effects?
|
gene expression
|
|
|
|
ARCHEA ONLY. have reverse gyrase which...
|
introduces positive supercoils.
|
|
|
|
the total complement of genes in a cell or virus
|
genome
|
|
|
|
ALL/MOST/NO prokaryotes have only one chromosome
|
MOST (few exceptions in bacteria and Archea which have 2 chromosomes)
|
|
|
|
ALL/MOST/NO prokaryotes have linear chromosomes
|
Most do not have linear chromosomes but a few do.
|
|
|
|
Nonchromosomal genetic elements include (4)
|
viruses, plasmids, organellar genomes, and transposable elements.
|
|
|
|
Viruses contain what kind of genetic material
|
Either SSDNA, SSRNA, DSRNA, DSDNA
|
|
|
|
Plasmids are
|
genetic elements that live inside of a prokaryotic cell and yet Replicate seperately.
|
|
|
|
Plasmids are both linear or circulae and found in what kind of cells...
|
Both prokaryotes and eukaryotes.
|
|
|
|
What is the difference between a chromosome and a plasmid?
|
Plasmids are extra chomosomal material. they do not encode for "necessary" calls.
|
|
|
|
ORganellar genetic material
|
Mitochondrion and Chloroplasts have their own genetic material believed to have evolutionarily been seperate organisms.
|
|
|
|
Transposable elements
|
molecules of DNA that can move from one site on a choromosome to another
|
|
|
|
DNA replication is said to be semiconservative meaning?
|
During replication, the strands are seperated and a complimentary strand is made of each original strand. thus every product is 1/2 new 1/2 old.
|
|
|
|
The addition of nucleotides to a replicating chain, must be to a
|
3' OH
|
|
|
|
DNA replication always precedes from
|
5' to 3'
|
|
|
|
Before a new chain can be started this nucleic acid molecule is necessary
|
Primer
|
|
|
|
Primer is of DNA/RNA
|
RNA, LATER IT WILL BE REPLACED BY DNA
|
|
|
|
The enzyme required to open the DNA to be replicated
|
Helicase
|
|
|
|
Origin of replication
|
the center of DNA synthesis fromwhich replication begins.
|
|
|
|
Once the DNA is opened by helicase ___________ enzyme keeps it unwound.
|
Single strand binding proteins
|
|
|
|
________ adds the nucleotides to the growing strain.
|
DNA Polymerase III
|
|
|
|
ON THE GROWING STRAND OF DNA TOWARD ORIGIN OF REPLICATION from 5' to 3' is the _________ strand
|
Leading
|
|
|
|
ON THE GROWING STRAND OF DNA TOWARD ORIGIN OF REPLICATION from 3' to 5' is the _________ strand
|
Lagging
|
|
|
|
Since nnucleotides are added only to 3' OH the lagging strand is replicated in fragments called
|
Okasaki
|
|
|
|
Next ____________ (enzyme) has exonuclease ability and proof reads the DNA as well as puts DNA in the place of primers
|
DNA Polymerase I
|
|
|
|
in bacteria since chromosome is circular, replication is
|
bidirectional
|
|
|
|
All the proteins used in this replication process work at the same time as a large unit knwon as a
|
replisome
|
|
|
|
How does Polymerase I and III know what nucleotides to remove?
|
The mismatch does not allow for the correct bonding. this however is DIFFERENT from the exonuclease activity that removes the RNA primer.
|
|
|
|
_________ recognize certain patterns of DNA and can excise fragments
|
Restriction enzymes.
|
|
|
|
What pattern do restriction enzymes typically notice
|
palindromic
|
|
|
|
What kind of break to restriction enzymes typically make?
|
Double stranded breaks leaving "Sticky ends"
|
|
|
|
What is the main function of restriction enzymes in prokaryotes?
|
It is their "immune system" by cutting out foreign genetic material.
|
|
|
|
How does DNA save itself from its own restriction enzyme?
|
it modifies its patterns of recognition by methylation.
|
|
|
|
Gel electrophoresis
|
the process of running genetic material on an electric plate.
|
|
|
|
Probes
|
molecules of DNA or RNA usually dyed to fluorsce or radioactive when then find the gene of interest.
|
|
|
|
Sequencing
|
the process of determining the exact order of nucleotides in a DNA segment.
|
|
|
|
PCR
|
the purpose is to multiply DNA molecules.
|
|
|
|
PCR takes advantage of high temps breaking the hydrogen bonds and then annealing together after replication, but how do DNA polymerase enzyme not denature?
|
Because the DNA polymerase used belongs to a thermophillic bacteria.
|
|
|
|
Diff between DNA and RNA
|
RNA contains ribose
DNA contains deoxyribose RNA contains U instead of T RNA is not double stranded. RNA has 2 OH groups DNA 1. |
|
|
|
mRNA
tRNA rRNA |
all are products of the transcription of DNA.
|
|
|
|
Common secondary structure of RNA
|
STEM LOOPS
|
|
|
|
Stem loops make what functional
|
tRNA
|
|
|
|
Transcription is carried out by
|
RNA polymerase (DNA template)
|
|
|
|
Unlike DNA polymerase what can RNA polymerase do...
|
Add nucleotides to a growing strand with no RNA primer.
|
|
|
|
In order for an RNA chain to begin correctly, they must first recognize this..
|
promoter
|
|
|
|
How long does the sigma factor and RNA polymerase complex remain in transcription.
|
Once a short stretch of RNA is made sigma complex dissociates; thus it seems to help recognize promoters.
|
|
|
|
How much transcription takes place in comparison to replication?
|
Very little. Alot less is transcribed sometimes only 1 gene.
|
|
|
|
A single organism has how many sigma factor
|
Several. the different allow RNA polymerase to recognize several promoter sequences.
|
|
|
|
RNA Polymerase I
RNA Polymerase II RNA Polymerase III |
I- synthesize most rRNA
II- synthesize all the mRNA (Transcription) III-synthesize tRNA |
|
|
|
TATA box
|
common in eukaryotes and Archea to highlight the promoter
|
|
|
|
Termination signal of transcription in prokaryotes
(Rho-independent) |
When RNA polymerase comes across a segment of inverted repeats it can form a stem loop followed by uradine runs.
|
|
|
|
Rho-dependent termination of transcription
|
Rho binds to mRNA and runs up the mRNA toward the DNA as soon as there is a pause (b/c mRNA reached a rho dependent determination site) mRNA breaks off.
|
|
|
|
Most genes encode proteins those that dont encode..
|
RNA's that are not translated such as rRNA and tRNA.
|
|
|
|
Unlike mRNA, rRNA and tRNA are
|
Stable
|
|
|
|
In PROKARYOTIC CELLS genes coding for similar enzymes are
|
often clusted together
|
|
|
|
Transcription of such clustered genes is known as ___________________ mRNA.
|
polycistronic
|
|
|
|
How many possible codons are there?
|
64 (4 bases in a series of 3)
|
|
|
|
there are 21 amino acids and 64 codons what are the rest "Coding for"?
|
Several amino acids have 2 codons that code for one amino acid (never does 1 codon code for different amino acids) Also there are start and stop codons to initiate and terminate translation.
|
|
|
|
Anticodon
|
the complimentary base pairings of the particular amino acid. recall that the codon (on mRNA) is the determinant of the A.A NOT THE ANTICODON.
|
|
|
|
Some tRNA's can recognize more than one codon, this is because
|
Wobble in structure. the third base does not have to match.
|
|
|
|
In order for mRNA to be translated it must contain an OPEN READING FRAME meaning
|
a start codon, followed by some number of codons and then a stop codon in the same reading frame as the start codon.
|
|
|
|
the genetic code is
|
universal
|
|
|
|
Codon biases
|
Codon biases exist- not all codons are used equally and this changes from organism to organism
|
|
|
|
tRNA and its specific amino acid are brought together by THIS enzyme that assures the right pairing
|
Aminoacyl-tRNA sythetases
|
|
|
|
the final and mature tRNA molecule is folded back on itself into what structure? Why?
|
Stem loop; this is how it is functional.
|
|
|
|
the site of protein synthesis
|
Ribosomes
|
|
|
|
the three steps of translation
|
initiation, elongation termination
|
|
|
|
initiation complex consists of
|
ribosome unit, mRNA, initiation proteins, GTPand a larger ribosomal subunit.
|
in translation
|
|
|
Just before the initiation codon on the mRNA is a 3-9 base nucleotide that aides in binding the mRNA and ribosomal subunits called:
|
Shine-Dalgarno sequence
|
|
|
|
Start codon in prokaryotes, eukaryotes and Archea
|
AUG= Formethionine (bactiera)
AUG= methionine (others) also GUG is the 2nd most common start codon. |
|
|
|
A site
P site E site |
Acceptor site
Peptide site Exit site |
|
|
|
A site
|
where new charged tRNA brought the next amino acid in the sequence of polypeptides.
|
|
|
|
P site
|
This is where the amino acid is actually tacked on to the growing peptide chain.
|
|
|
|
E site
|
this is where the uncharged tRNA leaves to cycle through again. of course the amino acid stays behind on the peptide.
|
|
|
|
Several ribosomes can translate one mRNA this complex is called
|
polysome (test question: consists of mRNA and ribosomes.)
|
|
|
|
Termination of translation
|
occurs when a nonsense codon is reached. No tRNA or Amino acid for that matter bind to the nonsense codon of mRNA.
|
|
|
|
A large number of antibiotics are known to inhibit protein synthesis by
|
interacting with ribosomes and rRNA specifically.
|
|
|
|
Upon completion of translation most proteins fold into confirmation alone but some require a protein called
|
Molecular chaperone
|
|
|
|
Collectively transcription and translation are known as
|
gene expression
|
|
|
|
2 levels of regulation
|
One controls ACTIVITY (post translational) and the other AMOUNT (transcription or translation).
|
|
|
|
Regulation of Enzyme activity: Feedback inhibition
|
Control of enzymatic activity to regulate biosynthetic processes. Where later products can inhibit the pathway at the beginning.
|
|
|
|
How does a final product stop a starting product?
|
The later product terminates the entire pathway by binding to its enzyme at an allosteric site and inhibiting it from forming the first intermediate in the pathway.
|
interupts the pathway how; enzymatically?
|
|
|
Different proteins that catalyze the same reaction. These diminish the effects of a pathway BUT DO NOT STOP it (like above)
|
Isoenzymes
|
|
|
|
Covalent modification of enzymes
|
where enzymes are regulated by modifying the enzyme and changing its shape
|
|
|
|
When there is extra genetic information they are removed before translation (post transcription) BUT if some remain it is removed after translation (gene regulation)in a process called
|
protein splicing
|
|
|
|
For transcription to occur, RNA polymerase must first recognize a
|
promoter and bind to it.
|
|
|
|
Histones are good examples of gene regulation (nonspecific)
|
because they are positively charged they bind to the negatively charged DNA. if tightly wound transcription cant occur.
|
|
|
|
Negative control of transcription
|
regulatory mechanisms that STOP transcription.
|
|
|
|
Recall that most of gene regulation occurs at
|
Transcription
|
|
|
|
Enzyme repression
|
Where the enzyme is not made until it is ABSENT. highly specific. Anabolic enzymes
|
|
|
|
Enzyme induction
|
an enzyme is made only when its subsetrate is PRESENT. Catabolic enzymes.
|
|
|
|
The enzyme that initiates induction is _________. The enzyme that represses enzyme synthesis is ___________. they are a class of _______.
|
Inducers; corepressor; effectors
|
|
|
|
Operons are a class of
|
Enzyme REPRESSION
|
|
|
|
An operon is
|
a cluster of genes arranged in a linear and consecutive fashion and whose expression is under the control of a single operator
|
|
|
|
Positive control of transcription
|
this is where a regulator protein ACTIVATES the binding of RNA polymerase.
|
|
|
|
UNDER POSITIVE CONTROL, transcription requires the activity of
|
an activator protein
|
|
|
|
The maltose activator protein cannot bind to the mRNA to begin translation until
|
maltose (effector) binds to the activator protein in the correct orientation to make the activator functional. recall this is all to allow transcription which is off (default)
|
|
|
|
Repressors bind to the operator; while activators bind to
|
activator-binding site
|
|
|
|
Diff between activators and repressors. Recall both STOP translation (Negative controls)
|
Operon is when "on is default" and the corepressor binding to the repressor protein (makes the correct conformation) for binding at the operator to BLOCK Transcription. (Arganine)
Induction: is when "off is default" or the repressor is in place. Then the inducer binds to the repressor and causes it to come off and Transcription begins. (Lac) |
|
|
|
Negative vs Positive control
|
Negative (above)
Positive- An activator protein is bound by an inducer and only then can it bind to the DNA to allow RNA Polymerase to begin transcription. (Maltose~ Also used for catabolic rxn) |
|
|
|
What is the purpose of the activator protein
|
when bound to DNA it helps the RNA polymerase recognize the promoter and begin transcription.
|
|
|
|
When an activator protein controls more than one operon it is known as a
|
regulon (this occurs because the genes required for a product are on different operons and so they come under the control of one activator protein).
|
|
|
|
Global control systems
|
regulatory mechanisms that respond to environmental signals by regulating expression of many different genes.
|
|
|
|
Catabolite repression
|
he synthesis of a variety of unrelated, primarily catabolic, enzymes are repressed when cells are grown in a medium containing glucose. Thus insuring glucose is used first (the best available carbon and energy source).
|
|
|
|
How does catabolite repression work?
|
CAP (activator protein) must be bound with cyclic AMP to bind to DNA and begin more transcription of lactose. BUT glucose (a better carbon source) binds to cAMP not allowing it to be transcribed at the same time it pumps cAMP out of the cell
|
|
|
|
Why is catobilite repression considered global?
|
because the presence of glucose, catalite repression prevents expression of all other catabolic operons.
|
|
|
|
Other examples of global regulation
|
Stringent response, alarmones, alternative sigma factors, heat shock response, cold shock and Quorum sensing.
|
|
|
|
Alternative sigma factors
|
global control by controlling the concentration of sigma factors. recall in order for transcription to begin a specific sigma factor is needed. thus controling its concentration u can control transcription of many genes requiring that 1 sigma factor.
|
|
|
|
Heat and cold shock proteins
|
Assist the cell recover from stress. these induce reactions when stressed or by heat or cold.
|
|
|
|
Quorum sensing
|
A method whereby a prokaryote recieve "signal" the presence in their surrounding of other cells of the SAME SPECIES. (for example to kill off other bacteria (Gram -) determine populations around them and then release toxin)
|
|
|
|
Attenuation
|
control systems that do not employ regulatory proteins to control transcription
|
|
|
|
Attenuation of the tryptophan operon
|
in addition to the promoter and operator on the DNA, there is a leader sequence that later encodes a polypeptide that will stop transcription.
|
|
|
|
When tryptophan is rich in the cell the leader peptde (will/will) not be synthesized
|
Will
|
what does the leader sequence do? stops transcription.
|
|
|
Mechanism of Attenuation
|
Attenuation occurs (transcription stops) because a portion of newly formed mRNA fold into a unique stem loop that causes cessation (3-4 stem loop) NOT 2-3.
|
|
|
|
Attenuation has been shown to control the biosynthesis of
|
Histidine (very detailed in notes)
|
|
|
|
Many times external signals (like in quorom sensing) are not directly sent to the regulatory protein but instead to a sensor that transmits the signal to the rest of the regulatory machinery this process is knwon as:
|
Signal transduction.
|
|
|
|
What 2 proteins are required for signal transduction and how does it work?
|
Sensory kinase and response regulator protein.
Kinases are proteins in the cell membrane that when they recieve information phosphorylate themselves to transduce messgaes. the phosphoro group is passed to a response regulator protein to effect transcription (usually block) |
|
|
|
Bacterial strains that grow in minimal media without added growth factors are
|
Prototropha
|
|
|
|
Genetic crossover is only possible for linear DNA (True/False)
|
False
|
|
|
|
DNA rearrangements involving crossover between homologus DNA sequence is called
|
Recombination
|
|
|
|
Ames' test is used to identify mutagens
|
True
|
|
|
|
Mutation
|
is an INHERITED change in the nucleotide base sequence of that genome.
|
|
|
|
Genetic recombination
|
the process by which genes contained in two seperate genomes are brought together in one molecule.
|
|
|
|
A mutatnt by definition differs from its parental strain in that ____________
|
its genotype is altered whether or not its phenotype shows this.
|
|
|
|
Selection
|
allows the isolation of a single mutatnt from a populatopm containing millions or billions of organisms.
|
|
|
|
In "NON SELECTIBLE MUTATION", how are mutatants picked out?
|
by careful screening of those that "look different" (this is because the mutant doesnt allow the organisms to live and the others die. F.e: a pigment.
|
|
|
|
Nutritionally defective organisms can be found by
|
replica plating- where plates of organisms (some with antibiotic mutation) are stamped onto another plate those that live acquired the mutation those that dont live didnt.
|
|
|
|
A mutant that has a nutritional requirement for growth is called...
|
an auxotroph
|
|
|
|
The parent generations that require no nutrittional requirements for growth (growth factors) are called:
|
Prototrophs
|
|
|
|
Pennicillin-selection method
|
A way to isolate the mutants that do not have a selectible mutation.
Pennicillin attacks only growing organisms so those that are growing are killed meanwhile those that cant grow live. |
|
|
|
Mutations can be either
|
spontaneous or induced
|
|
|
|
Spontaneous mutations can occur as a reult of...
|
exposures to radiation, radicals, or as most do DURING DNA REPLICATION.
|
|
|
|
Point mutations
|
mutatins in one base pair.
|
|
|
|
Silent mutations
|
mutations that change the codon but the incorrect codon still codes for the same amino acid and thus the primary structure is not ruined (common in the wobble position)
|
|
|
|
Missense mutation
|
Where a mutation causes a codon coding for a different amino acid than the one intended.
|
|
|
|
Nonsense mutation
|
This is when a mutation causes a termination of the translation of mRNA due to the wrong codon being a stop codon.
|
|
|
|
Frameshift mutation
|
because the genetic code is read in three bases= a codon. the insertion or deletion of one base changes the reading frame for all other bases this class of mutations are called frameshift mutations.
|
|
|
|
Translocation
|
the displacement of large amounts of DNA due to errors in recombination (in eukaryotes commonly relocate to another chromosome).
|
|
|
|
Inversion
|
when the orientation of a particular segment of DNA is reversed with respect to its surroundings.
|
|
|
|
Because naturally occuring mutations are rare, inducible mutations are more common they include:
|
radiation, contact with chemical, physical or biological reagents.
|
|
|
|
Nucleotide base analogs
|
Bases that mimic purines and pyrimidines and pair in their place causing faulty bonding and leading to higher rates of mutation.
|
|
|
|
Other chemical mutages lead to increase in faulty base pairings this is (not nucleotide base analogs)
|
Alkylation of molecules
|
|
|
|
How does UV light cause mutations
|
UV light causes pyrimidine dimers to form. the pyrimidines covalently bonded are misread during replication and leading to higher incidence of mutations.
|
|
|
|
Mutations arising from DNA repair
|
The SOS system this is where DNA is repaired. the problem some of the correcting is done "in the abscence of template instruction" meaning w/o base pairing leading to more mutations.
|
|
|
|
How does the Ames test test for mutations.
|
THe standard way to test for mutagenesis is to look for an increase in the rate of back mutation (Reversion) in auxotrophic strains of bacteria in the presence of the suspected mutagen.
|
|
|
|
recombination is
|
the physical exchange of genes between genetic elements.
|
|
|
|
Homologous recombination
|
results in genetic exchange betweeen homologous DNA sequences from two diffrent sources
|
|
|
|
The one on the right is positive and the one on the left is negative. Clumped in the middle means increased likelihood of mutation.
|
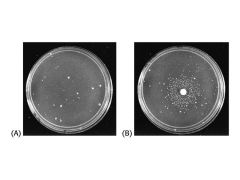
Which of the following is positive/negative for Ames test?
|
|
|
|
Genetic exchange in prokaryotes= Transformation
|
where DNA is released from one cell and is taken up by another. (Naked DNA)
|
|
|
|
Genetic exchange in prokaryotes= Conjugation
|
where DNA is passed from a F+ cell to a F- cell. Bacterial sex. Usually antibiotic resistance is passed.
|
|
|
|
Genetic exchange in prokaryotes= Transduction
|
Donor DNA transfer from bacteria to bacteria by having a virus as a vector.
|
|
|
|
In transformation before genetic material can be taken up host cells must be _______. How do we achieve this?
|
competent; CaCl2, Heat shock, or electric shock.
|
|
|
|
Genetic exchange in prokaryotes= tranasfection
|
where bacteria accept genetic material from a BACTERIAL VIRUS.
|
|
|
|
Plasmids are genetic elements that
|
replicate independently of the host chromosome and exist in the cell simply as free DNA material (usually circular).
|
|
|
|
When a plasmid is transferred into a cell that already carreies another plasmid a common observation is...
|
incompatibility.
|
|
|
|
Episomes are
|
plasmids whose DNA has been incorporated into the host cell's DNA and comes under the control of the host cell.
|
|
|
|
Curing
|
removal of a plasmid from a host cell.
|
|
|
|
Cell to cell transfer of cells
|
Since some prokaryotes can take up free DNA, it is possible that lysing a cell is a form of communication to find a new host.
|
|
|
|
The main mechanism of cell to cell transfer of plasmids is
|
conjugation (prokaryotic sex)
|
|
|
|
Transposition
|
the process by which genes move from one place to another. "Jumping genes"
|
|
|
|
Transposition is not random, it is linked to what special genetic elements
|
transposable elements
|
|
|
|
Three types of transposable elements in bacteria
|
1.) insertion
2.) transposons 3.) special viruses |
|
|
|
The enzyme necessary for transposition
|
Transposase
|
|
|
|
Besides transposase what else is coomly seen in transposition
|
short inverted terminal repeats so that insertion and complimentation have sticky ends to seal DNA.
|
|
|
|
Insertion sequences
|
the simplest of transposition. this is where a transposable element is cut out and put in somewhere else only with the genes required for the move.
|
|
|
|
Transposons
|
are larger than insertion sequences and carry other genes some of them conferring important quantities.
|
|
|
|
Conjugative transposons
|
those transposons that can also be passed from organism to organism through conjugation.
|
|
|
|
when the transposable element is excised from one location on the chromosome and becomes reinserted in a second location
|
Conservative transposition (1 copy to 1 copy)
|
|
|
|
when a transposoble element makes a new copy and the copy is inserted in a new location
|
replicative transposition (1 copy to 2 copies or more)
|
|
|
|
Transposition is like recombination except that
|
it is not between homologous pairs.
|
|
|
|
Insertions of transposons within gene cause
|
frameshift mutations
|
|
|
|
transposons that can capture and express genes from other sources.
|
Integrons
|
|
|
|
What is the difference between an integron and a transposon?
|
Transposons add themselves into the genetic material RANDOMLY.
Interons are highly specific in their binding, often binding to plasmids. |
|
|
|
How are integrons so precise in their binding?
|
they have a protein called integrase needed for site-specific recombination.
|
|
|
|
3 essentials of molecular cloning
|
1.) Isolation and fragmentation of of source DNA.
2.) Joining the DNA fragments to a cloning vector with DNA ligase. 3.) Introduction and maintenance of the cloned DNA in a host organism. |
|
|
|
Site-directed mutagenesis
|
Unlike most mutations that occur at random. Site directed litterally occurs at a particular site.
|
|
|
|
Backbone of DNA consists of... (how are they bonded)
|
Alternating segments of Phosphate, sugar and base. the phosphate and sugar are held together through a covalent phosphodiester bond. Wheras the dsDNA is held together by hydrogen bonding.
|
|
|
|
Know how to calculate the % of bases.
F.e: you are told that a segment of DNA has A= 13%, what percentage of DNA is made of Guanine. |
A= 13%
T= 13% G + C = 100-26= 74%/2 = 37% of the DNA is thus Guanine. |
|
|
|
what is the theta structure?
|
During DNA replication when there are 2 origins of replication (forks) the seperartion of DNA strands from the original strand is called a theta structure.
|
|
|
|
This enzyme unwinds duplex DNA
|
Helicase
|
|
|
|
this protein binds to and stabilizes ssDNA
|
ss binding proteins
|
|
|
|
Using a DNA template this enzyme synthesizes a short RNA segement to begin replication.
|
Primer
|
|
|
|
this molecule has the ability to replicate and proofread but NOT to remove primers.
|
DNA polymerase III
|
|
|
|
This molecule can synthesize DNA from a DNA template and removes RNA primer.
|
DNA polymerase I
|
|
|
|
What activity allows polymerase I and III to proofread and specifically correct a base as well as remove primer (I only)?
|
Exonuclease activity
|
|
|
|
Telomeres are (always, sometimes, mostly, never) found in (prokaryotes, eukaryotes, Archea)
|
Only in eukaryotes.
|
|
|
|
Why dont bacterial chromosomes have telomers or the need for telomerase?
|
because bacterial chromosomes are USUALLY circular and there is no end to replication.
|
|
|
|
telomerase is a product of
|
a protein and an RNA molecule
|
|
|
|
Stem loop structures occur only in
|
RNA
|
|
|
|
How many bonds between the base pairings of G and C vs A and T
|
3 ; 2
|
|
|
|
Why is the stemloop so important? (hint: in translation)
|
Makes the structure of tRNA functional.
Also it is a terminator sequence for transcription (rho-dependent) |
|
|
|
How many genes in a prokaryotes, eukaryote, virus?
|
Virus- hundreds
Prokaryotes- thousands Eukaryotes- tens of thousands |
|
|
|
During transcription how many strands are changed from DNA to mRNA? Of which strand.
|
During mRNA synthesis in transcription one strand of DNA is used to code for mRNA and is thus called the coding strand. the other is called the complimentary strand.
Lastly the mRNA is an EXACT copy of replicated DNA. ( |
|
|
|
Distinction between operon and polycistronic message
|
Operon- two or more genes transcribed together under the operation of the operator.
Meanwhile polycistronic DNA is a single RNA that represents more than one gene. |
|
|
|
In eukaryotes only, after transcription, modifications occur what are they, why are they done and where are they done.
|
A Guanine cap and a poly A tail is added. to protect the molecule from degradation as it leaves the nucleus into the cytoplasm.
|
|
|
|
besides cap and tail what else is done post transcriptionally?
|
splicing of introns and ligating exons back together.
|
|
|
|
Eukaryotes do not have
Prokaryotic genes very rarely have |
operons
introns |
|
|
|
Where RNA polymerase binds on the DNA for transcription to begin
|
Promoter (upstream from start of gene)
|
|
|
|
the enzyme that synthesizes mRNA from DNA.
|
RNA Polymerase II
|
|
|
|
Two conserved regions or promoter regions are at
|
-35 and -10 (TATA box
|
how many base pairs from the promoter and what specific region?
|
|
|
RHO independent transcription termination
|
this is where the DNA encoding an RNA that forms a stem loop followed by a run of U's is terminated from further transcription.
|
|
|
|
RHO dependent transcription termination
|
A DNA site where RNA polymerases pauses ans transcription is terminated by Rho protein.
|
|
|
|
Sigma factors usage?
|
Sigma factors are needed for promoter binding (which in turn is required for polymerase II binding). Once promoter is bound to DNA sigma factor dissociate.
|
|
|
|
Core enzyme vs. holoenzyme
|
Core enzyme has no sigma factor.
Holoenzyme has several sigma factors. |
|
|
|
most gene regulation occurs in
|
Transcription
|
|
|
|
what is the 16sRNA
|
the site of Shine Dalgarno sequence of mRNA (promoter for ribosome binding)
|
|
|
|
Prokaryotes and eukaryotes have different # subunits in ribosomes
|
prokaryotes- 50s and 30s
eukaryotes- 40s and 60s |
|
|
|
Polysome
|
mRNA with several translating ribosomes attached.
|
|
|
|
Replisome vs. Polysome
|
Replisome- when replicating the complex that forms between DNA polymerase III, helicase and DNA primase of the leading and lagging strand.
Polysome- when RNA polymerase II is transcribing DNA there may be a few translating ribosomes as well. the complex is called a polysome. |
|
|
|
Wobble hypothesis states that the 3 nucleotide does not always have to be exact this gives rise to what...
|
codon families where one of the nucleotides misplaced or incorrect can still code for the same amino acid.
|
|
|
|
how to perform Ames' test
|
1.) spread minimal plate of Histidine - cells.
2.) soak filter disk with test compound and place on plate 3.) incubate plates and examine. |
|
|
|
Why are restriction ezymes used (only in microbes)?
|
to protect bacteria from viruses. in essence the bacterial immune system.
|
|
|
|
Basic steps of DNA cloning. (5)
|
1.) isolate DNA of interest
2.) Digest DNA and vector with the same restriction enzyme 3.) ligate the source DNA to the vector. 4.) introduce into a host 5.) identify the clone of interest. |
|
|
|
DNA libraries
|
a large number of clones representing the entire genome of an organism.
|
|
|
|
important features of a clone vector
|
Selectible markers (antibiotics f.e to test for mutation)
means of replicating unique restriction sites acceptable insert sizes. |
|
|
|
Phage lambda
|
a viral cloning vector where 1/3 of its DNA can be replaced.
|
|
|
|
Cosmids
|
plasmisds with cos sites for in vitro packaging into lambda capsids (?)
|
|
|
|
Site directed mutagenesis
|
mutation at a direct source by recombination of homologous chromosomes.
|
|
|
|
Clone sizes
|
Plasmids allow 10 kbps
Cosmids ~ 45 kbps YACS up to 800kbps |
|
|
|
Repressor proteins
|
Decrease transcription by interfering with RNA polymerase binding to the DNA
|
|
|
|
Activator proteins
|
Increase transcription by activating the site where which RNA polymerase binds to the DNA
|
|
|
|
Binding sites for repressor and activator proteins are found
|
within palindromic repeats and close to promoters.
|
|
|
|
EFFECTORS have what structure
|
DIMERS; they change the DNA binding affinity of regulatory proteins for the binding site.
|
|
|
|
Gene regulation
|
Triptophan operon
Always On until repressed Common in biosynthetic or anabolic reactions |
|
|
|
Gene induction
|
Lactose operon
Always off until required Common in catabolic reactions |
|
|
|
Catabolite repression
|
A global regulatory system that allows glucose to be consumed in preference of other carbon sources.
|
|
|
|
Recall that the lactose operon is under a different regulation (not only lac operon)
|
CAP/cAMP cycles also regulate lactose enzymes. glucose however limits the amount of cAMP and exports it out of the cell (catabolite repression)
|
|
|
|
Sensor kinase
|
Integral membrane proteins that sense environmental changes and through phosphorylation send cascading messages.
|
|
|
|
Attenuation
|
controlling gene expression by control of transcriptional termination
|
|
|
|
Histadine Attenuation of the his operon
|
When histadine is plentiful stem loop occurs between 1 & 2 and 3 & 4 with tail of U's ending transcription (rho-independent)
when however it is low the stem loop structure has a loop between 2 and 3. |
|
|
|
messenger RNA contains
|
Ribosome binding site
|
|
|
|
Telomerase has what template
|
RNA
|
|
|
|
Small molecules from the environment (effectors) regulate gene expression by binding to
|
regulatory proteins and changing their affinity for binding with DNA and beginning transcription.
|
|
|
|
transcriptional repressors bind to
|
Operator sites not promoters.
|
|
|
|
the purpose of sgma factor is to promote
|
RNA polymerases binding to the DNA to initiate transcription
|
|
|
|
two daughter cells are most likely to inherit what from parental cells
|
a change in a nucleotide in DNA
|
|
|
|
What is the function of the enzyme reverse transcriptase
|
to make a molecule of DNA from RNA.
|
|

