![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
83 Cards in this Set
- Front
- Back
|
General Principal Reminders
|
Bacteria are Prokaryotes and are Haploid (heritable unit is a single copy of the genetic material).
They Typically divide via binarry fission into genetically identical offspring. Genetic Diversity Arises via Mutation and Genetic Exchange(recombination) with other populations. |
|
|
Mutation
|
Heritable changes in the nucleotide sequence of the bacterial genome (chromosome). Usually they are detected phenotypically rather then vis sequencing genotypes. Note that in a haploid organism the lack of a backup allele causes all mutations to be apparant.
Mutation rate is normally about 1 X 10^-6, (1 X 10^-12 for having 2 mutations simulatneously, being the reasoning for combined antibiotic therpy). Ex...The mutation rate for rifampacin resistance in 1x10^-7 or 08 (as only the B rna polymerase subunit mutation gives immunity) however there may be 1x10^-7 organisms in a TB lesion of the lung. |
|
|
Rate of Genetic Change
|
Occurs considerable faster than the mutation rate due to transfer of genes between bacterial populations via transformation, transduction and conjugation (all mediated via homologous recombination between a donor/exogenote and recippient/endogenote).
Note that one hallmark of recombianton in E-coli is the requirement of the RecA protein (which is required elsewhere as well. non-homologous recombination is also possible via RecA independant mechanisms including transposition, phage lysogeny, and phase variation. |
|
|
Are there mutants of bacteria that are deficient in genetic recombination?
|
Ans. to ques. 1, Yes. Any gene can mutate, so can the recA gene (and other genes that are
needed for recombination). During normal growth, homologous recombination is used by the bacteria to restart stalled DNA replication forks, which can result from replication error. recA mutants cannot do that, and so are sick. |
|
|
Transformation
|
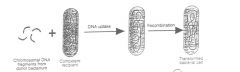
a process whereby free, soluble, naked DNA from one bacterium (the donor) gets
into another (the recipient). - usually, there need be no polarity. (In other words, the recipient can also act as donor for the DNA and the donor can act as a recipient.) The process id DNASE susceptable. Competence - the state of a bacterium that allows it to take up DNA. In some species this occurs only in certain growth conditions. In some species competence is because of a protein competence factor on the surface of the recipient. Gram-positive bacteria (e.g. Streptococcus pneumonia and Bacillus subtilis) - only a single strand of DNA (ssDNA) enters the recipient, the other strand is digested at the surface before the entry process. Gram-negative bacteria (e.g. Haemophilus influenzae, Neisseria gonorrhoeae [ability to transform --> pathogenicity]). Firstly, double-stranded DNA is taken up, although only one strand integrates into the recipient chromosome. Secondly, determinants on the surface of each species recognize a signature sequence in the DNA, so that DNA from other species is largely excluded. |
|
|
Transformation Mapping
|
Two or a few genes can be co-transformed as a single piece of DNA, showing that they are
linked. Thus, if gene A is found to be linked to B (i.e. it is co-transformed), and B to C, but C is not co-transformed with A, then the linkage map is A-B-C. Remember that at most 2% of the donor chromosome is transferred in such crosses, so that linkage is highly significant. Percent cotransfer can be used to give a crude measure of map distance. Transformation of E. coli (where competence requires Ca++, probably for making transient holes in the cell membrane) is one of the major technical requirements for recombinant DNA experiments. |
|
|
Transduction
|
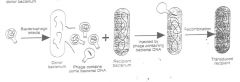
A process in which donor bacterial DNA is incorporated into an empty phage head
(by a rare mistake in packaging of the phage head) when phage particles are being assembled inside the infected bacterium. The phage particle can now transfer the bacterial DNA to a recipient. Generalized transduction - As with transformation, any fragment of donor chromosome can be transferred. Specialized Transduction- restricted to special parts of the chromosome. Transduction is found in a variety of gram-positive and gram-negative bacteria. The only requirement is the availability of a suitable phage for the bacterial species. Like transformation, transduction can be used to map genes on the chromosome of a species, because linked genes are co-transduced. |
|
|
Given two bacterial strains - A, which requires arginine and is resistant to
streptomycin, and B, which is a prototroph (no nutritional requirement), but is susceptible to streptomycin. Both are grown separately in a nutrient broth and are then mixed and incubated for an additional hour or so. Now you streak on an agar medium lacking arginine but containing streptomycin, and you subsequently observe growth of colonies. Given the choice of transformation or transduction, what is the simplest way of eliminating transformation as the process of genetic transfer in a repeat experiment? |
Mix A and B in the presence of deoxyribonuclease and as a control mix
them in its absence. If no recombinants can be found from the DNAse-treated mixture, but are obtained in the absence of DNAse, then the process was transformation. Transduction is resistant to DNAse. |
|
|
Conjugation
|
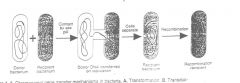
bacterial genetic exchange involving sexual polarity. There are distinct donor and recipient strains within a species.
Pilus - a structure, made of protein subunits, that is necessary for conjugation. It appears to act as a grappling hook to establish cell-to-cell contact, which results in DNA transfer. Contact - via pilus and/or closer intimate contact, which might be the result of the pilus retracting inside the donor and pulling the recipient into close contact with the donor. The requirement for cell-to-cell contact distinguishes conjugation from transformation and transduction. Conjugation often results in transfer of more DNA (genes) than transduction or transformation. As much as one third of the chromosome can be transferred by Hfr-mediated conjugation, but in practice no more than one third is transferred. In comparison, rarely more than 2% of the chromosome is transferred in transformation and transduction. Conjugation Between Strains of E. coli K12 Depends on the F (Fertility) Factor |
|
|
F Factor
|
F factor is a plasmid (in older literature it is sometimes referred to as an episome). It is
double-stranded, circular DNA. It is about 1/50 the size of a chromosome. The F plasmid is its own replicon when it is extrachromosomal. The F factor is unusual among plasmids, in that it has a high affinity for inserting into the host chromosome, becoming part of that replicon. F can integrate into about 25 different sites of the chromosome because it and the chromosome have short homologous sequences. |
|
|
F+ x F- Mating
|
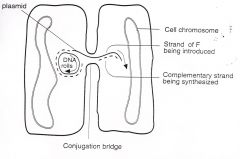
pilus makes contact with recipient, triggers F replication and transfer of
one DNA strand of F to the recipient where that strand is replicated. The strand left behind in the donor is also replicated. The entire sex factor (F) goes over, converting the recipient to a donor. In a mixture of donors and recipients, the transfer is rapid and widespread; the entire population becomes F+. This is sometimes referred to as an infectious, or epidemic, transfer of the F plasmid. (Other, similar plasmids are also capable of rapid and widespread dissemination; some carry drug-resistance determinants.) Occasionally the entire F factor integrates into the bacterial chromosome to generate an Hfr strain |
|
|
Replicon
|
A replicon is a “unit of DNA replication”, having its own
origin of replication. Bacterial chromosomes and plasmids are replicons. Any piece of DNA inserted into a replicon will replicate as part of that replicon |
|
|
Hfr x F- Mating
|
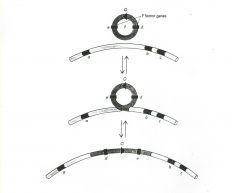
The F factor is incorporated into the chromosome in Hfr strains (Hfr = High Frequency Recombination). As such, it becomes a part of the chromosome replicon. However, when the pilus attaches to a recipient, the chromosome behaves as if it is
a part of the F replicon. The chromosome is transferred to the recipient in an oriented linear fashion. Part of the F factor DNA is destined to go over last. Usually, if not always, the transfer spontaneously stops (picture it as though the connecting bridge is broken) and, at most, 1/3 of the chromosome is transferred. As a consequence the recipient remains a recipient because part of the F factor is never transferred to the recipient. There is a high frequency of recombination between the introduced portion of the donor chromosome and the recipient chromosome. However, Hfr mating is not infectious. |
|
|
F' generation and mating
|
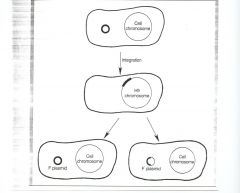
The F factor can integrate into the chromosome to give an Hfr. It can also excise from the chromosome. If excision is precise, it gives back the F factor. If excision is imprecise, it can
give back a modified F factor, called F’ (F-prime). There are three types of conjugation, depending on which of three interchangeable forms of the F factor is present in the donor. F’ <-> Hfr <-> F+, insertion of F factor into the chromosome and its excision from the chromosome are rare events (10^-6/generation), and so one can obtain fairly stable F+, F’ and Hfr strains. F’ x F- mating - if one, two or few chromosomal genes are excised along with the F factor when it is excised from the chromosome, those genes will be transferred with the F factor. Those genes are part of the F replicon. As in the F+ mating, the recipient becomes a donor upon receipt of F’. That is, F’ transfer is infectious (or epidemic). |
|
|
Interrupted Mating
|
Although the mating act spontaneously stops, one can
deliberately stop (interrupt) mating by vigorously shaking the mating population in a blender. Samples of the mixture are taken at time intervals, blended and then plated onto agar to select recipients (recombinants) with appropriate genetic markers. The rate of appearance of recombinants for different markers, gives the time of entry of the marker and its location on the chromosome. The conclusion from such experiments is that there is an orderly, linear linkage of genes and that the genetic map is circular. The genetic map of E. coli K12 was constructed using the processes of conjugation and generalized transduction and the complete DNA sequence of the E. coli genome is in excellent agreement with the genetic map. |
|
|
Conjugation of other Gram -ve's
|
F factor-mediated conjugation also occurs in Salmonella typhimurium. Conjugation in other species,
for example, Vibrio cholerae and Pseudomonas aeruginosa, is mediated by other sex factors. The latter mating systems have not been so well studied. |
|
|
Conjugation of Gram +ve's
|
The prototype system is that of Enterococcus faecalis. The mechanism of conjugation is very different from that of E. coli. The donor cells (plasmid-containing) are stimulated to produce an adhesin (note: there are no sex pili) when the donors sense a 7-residue peptide
pheromone (a hormone) produced by a recipient cell. This results in a clumping of donors and recipients, and subsequent transfer of DNA by cell-to-cell contact. Other gram-positive bacteria (as examples, members of the genera Bacillus, Staphylococcus, Clostridium) also have conjugative plasmids, but less is known about these mating systems. Of major medical importance is the fact that the conjugation systems can account for the transfer of plasmid-mediated antibiotic resistance and even toxin production. Conjugation is the major mechanism for the spread of antibiotic resistance in the Enterobacteriaceae - Escherichia, Salmonella, Shigella. |
|
|
Phenotypic Lag
|
This is the delay in phenotypic expression of a genotypic change. This is an
IMPORTANT factor in the spread of antibiotic resistance - either by mutation or by genetic exchange. There is a gap between the introduction of a gene (basically naked DNA) into an organism and the expression of the resistance that it determines. Phenotypic lag is one reason why it is important that there are no lapses in antibiotic therapy to be sure that the therapy removes a pathogen from the system. The actual timing of the lag is variable. |
|
|
DNA was extracted from a strain of Haemophilus influenzae resistant to
streptomycin (Str), erythromycin (Ery) and nalidixic acid (Nal). The DNA was mixed and incubated with a strain of H. influenzae susceptible to those antibiotics. After a short period of time, the mixture was plated on three different nutrient agar plates - one containing Str, another Ery, and the third Nal. After incubating the agar plates, about an equal number of colonies grew on all three antibiotic media. One hundred colonies from each plate were tested for resistance (or susceptibility) to the other two antibiotics. The results were as follows: Selected marker Num resistant Str Ery Nal Str 100 40 0 Ery 20 100 1 Nal 0 5 100 Which one of the following can be concluded? a. str and ery are most closely linked. b. ery and nal are most closely linked. c. nal and str are most closely linked. |
a. nal and str are not linked.
|
|
|
(c) Leu, Pro, Lac, and Gal, respectively.
|

The figure below illustrates the results of an interrupted mating experiment using
Hfr 1. Remember that usually only about 1/3 of the chromosome is transferred in an Hfr mating. Markers 1, 2, 3 and 4 are (a) Lac, Pro, Leu and Thr, respectively; (b) ArgE, Thr, Leu and Pro, respectively; (c) Leu, Pro, Lac, and Gal, respectively. |
|
|
If a membrane filter kept the donor and recipient bacteria apart, could gene transfer
occur by the process of conjugation? transformation? transduction? |
the membrane filter prevents conjugation because
conjugation requires direct contact between donor and recipient. It does not prevent transformation or transduction because DNA and phage can pass through the filter. |
|
|
Bacterial RNA Polymerase
|
Bacterial RNA polymerase consists of a core of four protein subunits, α2ββ’. A fifth protein
subunit, σ (sigma) binds to the core to give α2ββ’σ, which is called the holo enzyme. The holo enzyme then binds to a DNA sequence called the promoter and starts to transcribe the DNA. The promoter itself is located just upstream from the transcription start point, and transcription proceeds in the direction away from the promoter (hence we say the promoter is upstream). Note: The structure is significantly different from that of eukaryotic RNA polymerases. This difference is a good indicator to search for antibiotics. Bacterial RNA polymerase is the target of the rifamycin series of antibiotics (e.g., rifampicin: used in the treatment of tuberculosis and leprosy; and in meningococcal prophylaxis). Unfortunately mutation to rifampicin resistance does occur; the mutations are in the gene for the β subunit. |
|
|
σ factor
|
Core enzyme does not bind specifically to the promoter. It is the σ factor that confers
promoter recognition on the holo enzyme. The holo enzyme binds to the promoter region to form a closed complex. Then part of the DNA double helix unwinds to form an open complex. This initiates transcription. Note that transcription and Translation are coupled. There sre secerl sigma factors in an organism, and ones that direct strong transcription conform closely to a "promoter consensus Sequence" most often located either 10 or 35 Bp upstream of the transcription start site. |
|
|
Termination of Transcription in Bacteria
|
Transcription continues until a transcription termination signal is encountered. Terminators
are structures in the recently synthesized RNA. There are two types: Rho-dependent and Rho-independent. Rho-dependent terminators need a protein Rho for termination to take place. Rho-independent terminators can form hairpin structures followed by a run of U's. Note that translation hinders the terminator structures and can often prevent termination |
|
|
Regulation of Transcription
|
Most regulation of prokaryotic gene expression affects initiation of transcription. This includes both Negative and Positive controls
|
|
|
Negative Control
|
lac operon of E. Coli is the paradigm.
E. Coli is usually lac + (red colonies on McKinkey's Agar, while Lac - cannot ferment lactose and lower pH.). It includes 3 structural genes coding for a polycstronic mRNA. The product of lacZ is β-galactosidase; it catalyzes the hydrolysis of β-galactosides to metabolizable sugars, e.g., lactose → galactose + glucose. The permease (lacY product) catalyzes uptake of β-galactosides by E. coli. The role of the transacetylase (lacA product) is unclear. lacI is the structural gene for the Lac repressor (regulator). It is an allosteric protein, which has dual affinity for β-galactosides and for the O region. It is the repressor, which binds to the O region keeps the lac operon turned off when a β-galactoside is not present. This is an inducible enzyme system. O - the operator, is the "on-off" switch for transcription of the operon; there is no transcription (off) when the repressor protein is bound to the operator; there is transcription (on) when the inducer, in this case the β-galactoside, is present and binds to the repressor, altering its conformation so that it loses affinity for the operator. Note that lactose does not act as an inducer. P - promoter is where RNA polymerase binds. Note that the system can be altered...in the trp operon the Trp repressor only represses the trp operon in the presence of tryptophan. It does not repress when there is no tryptophan. |
|
|
Operon
|
a group of adjacent genes whose transcription is regulated from a single operator
region. Thus the lac operon includes Z, Y, A, and most authors also place O and P in the operon. The lacI gene is not part of the operon, and works equally well when it is moved to a different part of the chromosome. The lacI gene has its own promoter (not shown). |
|
|
Cis/Trans Acting Factors
|
TRANS-ACTING FACTORS AND CIS ELEMENTS are the fundamental components for the regulation of transcription. Expression of the lac operon on both the chromosome and the F’ factor (of a merodiploid) is
inducible. The LacI repressor protein is a trans acting factor. OC determines the expression of the lacZ,Y,A structural genes that are immediately adjacent to it, on the same DNA (e.g. the F factor), but does not determine the expression of lacZ,Y,A structural genes on some other DNA (e.g. the chromosome). In other words, the operator is a CIS element. |
|
|
Positive Control
|
The arabinose operon is the paradigm. The operon includes structural genes B, A, and D, as well as Apo C which is just upstream from the operator. The operon is only transcribed in the presence of arabinose and the protein product of the
araC gene. This is a positive control system - you need the regulator, AraC, to get expression. The AraC protein is often called the activator. Catabolite repression is another example of Positive Control. Glucose effect on E. Coli is the paradigm. When Glucose is low, cAMP lecels rise, allwing cAMP to bind to cAMP regulator protein (CRP) AKA Catabolite gene Activator Protein (CAP). The active complex is a positive and necessary regulator for the lac operon, so that then glucose falls transcription is released, but when glucose is high transcription is prevented even in the presence of lactose. |
|
|
How can you tell positive from negative control
|
Delete the regulatory gene: delete lacI or
trpR and you get constitutive expression of the corresponding operons (that means the operons are expressed all the time whether or not lactose is present or tryptophan is absent, respectively). Delete araC and you get no expression of araBAD whether arabinose is present or not. Note that AraC, LacI and TrpR are all trans-acting factors. |
|
|
Global Regulation
|
An organism effects major changes in metabolism in response to environmental changes. Examples:
Catabolite Repression - effecting lac, ara. and other operons for poor carbon source utilization. SOS Response - Genes for DNA repair and to block cell division are usually repressed via LexA. The RecA protein (also used for homologous recombinatoin) is activated as a result of DNA damage and causes auto-proteolosys of LexA disinhibiting the system. Alternate Sigma Factors - spore formation by Bacillus and Clostridium where there are specific necessary sigma factors with individual promoters. 2 component sensor-effector regulation - Responds to environmental changes via a histidine kinase (sensor) that autophosphorylates when activated. The activated complex then phosphorylates an effector, often a regulator of transcription. Often the state of these systems mediates virulence (in shigella flexneri, Staph Aureus, N. Gonorrhoeae, Salmonella typhmurium, and bordatella pertussis for instance). These systems have not been found in hihger eukaryotes, making them new antibiotic targets. |
|
|
Attenuation
|
A form of specific control of transcripion (ex. amino acid synthesis operons like his and trp of E. Coli). It functions because transcription and translation are coupled. In the leader region, there is a leader peptide that can only be completed in the presence of the amino acid in question (Histidine for his etc). Between the leader peptide and structural genes is an attenuator region which assumes a hairpin conformation. When the leader peptide is transcribed quickly (the AA is abundant) the conformation terminates transcription. When it stalls waiting for the tRNA the conformation differs such that transcription proceeds. Note that attenuation is the ONLY specific regulation of his, but trp is also regulated via repression).
|
|
|
DNA is double stranded, but only one strand of the lac operon is transcribed into
mRNA. How does the cell know which strand is correct? |
Ans. to ques. 2 is the promoter provides orientation
|
|
|
3: Look at the mRNA for the structural genes in Fig. 4. We need chain-terminating (or
stop) codons, UAG or UAA or UGA, to get a protein from a gene (one gene-one enzyme or protein). Therefore, terminating codons are needed: a) between the Z and Y messages, and between the Y and A messages b) between the Z and Y messages, the Y and A messages, and at the end of the A message |
b) between the Z and Y messages, the Y and A messages, and at the end of the A message
|
|
|
What does lacZ code for?
|
Ans. to ques. 4 is β-galactosidase. One of the few gene names you should know.
|
|
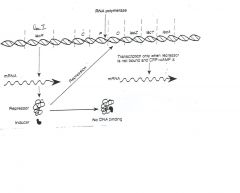
Deletion of which of the following could make E. coli
lactose-negative, i.e., loss of ability to ferment lactose: a) P b) Z or Y c) I d) a and b e) a and c f) b and c g) a, b and c |
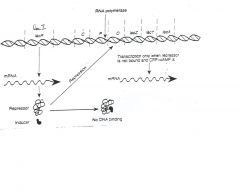
Ans. to ques. 1 is d. Deletion of I leads to constitutive expression; deletion of Y prevents lactose
uptake; deletion of Z prevents lactose metabolism. |
|
|
Plasmids (general Properties)
|
1. Covalently closed, double-stranded, circular DNA.
2. Not essential for bacterial growth under most conditions. 3. Replicate independent of the host chromosome, therefore they are replicons, and partition efficiently into both daughter cells at the time of bacterial cell division. (note that they often have thier own origen of replication and can be present with 1 copy/chromosome or many. Some can integrate into the chromosome (being then renamed episomes) 4. Recently plasmids are named thus pBR322... p=plasmid, BR=Lab Code, 322=lab number to pinpiont plasmid. Does not apply to older plasmids like "F" |
|
|
Plasmid mediated resistance
|
Many plasmids carry genes for antibiotic resistance. These plasmids are sometimes referred to as R plasmids or R factors. Often they carry several genes conferring resistance to several
different antibiotics. Many of the resistance determinants are parts of transposons contained within the plasmid. The plasmid (or transposon) introduces a whole new gene that determines the resistance. Plasmids cause resistance in a variety of ways: Notably by inactivating the antibiotic, by altering the target or by pumping the antibiotic out. For most gram-positive and gram-negative species, plasmids are the main source of clinically important antibiotic resistance. |
|
|
Examples of Plasmid-Mediated Mechanisms of Resistance to Antimicrobial Agents
|
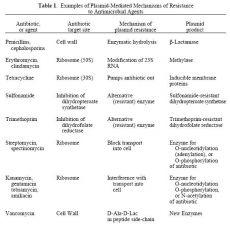
See Chart
|
|
|
Plasmid Mediated Pathogenicity
|
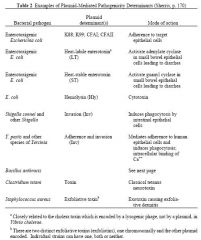
Some plasmids contribute to bacterial pathogenicity. They do so in different ways: notably toxin production, adherence to target cells, survival in host. The role of plasmids in pathogenicity has been worked out by "curing" bacteria of their
plasmids (i.e., causing the bacteria to lose their plasmids) in the laboratory. Curing can be caused by acridine dyes or UV light. The major virulence factors of B. anthracis are encoded by two large plasmids, pXO1 (exotoxin) and pXO2 (capsule). Some of the nastiest pathogens of all time are plasmid dependant. |
|
|
Bacteriocins
|
Many plasmids code for bacteriocins, which are secreted proteins that are lethal for cells of
the same or related species; the producing strain is immune. In other words, bacteria with such a plasmid kill off related bacteria that do not contain the plasmid. Bacteriocins are often named after the species producing them, such as colicins from E. coli, megacins from Bacillus megaterium. They kill in many ways: Nuclease Action (DNAse/RNAse) Ionophore (dissipates membrane potential) Note that the host range is very specific so that the bacteriocins can be sued to type pathogens, and thus identify the Index Case of an epidemic (such as Shigella causing bacillary dysentery or Pseudomonas Aeruginosa which is opportunistic with cystic fibrosis and burns). |
|
|
Plasmid Transfer
|
Plasmids are DNA and thus can be transferred through transductin, transformation or conjugation. Those using conjugation are called conjugative plasmids and ahve an origen of conjugation called oriT (different than the oriS origin of replication). Many nonconjugative plasmids can be mobilized by (hitch a ride with) a conjugative
plasmid and so be transferred from bacterium to bacterium. Thus an innocent plasmid such as F causing no known drug resistance can mobilize a small nonconjugative plasmid that does determine drug resistance. Note that all 3 methods are found in nature, but not all bacteria use all 3. Conjugation is probably the most effective. Also note that plasmids can be transferred between different species wtih varied levels of promiscuouity depending on the recippient. Factors that reduce plasmid establishment in a new species include: Restriction endonucleases Plasmid replication machinery does not work Inefficient transcription Translation requirements Codon usage GC content of DNA Also, genes can move from plasmid to plasmid/chromosome as part of a transposon |
|
|
Plasmid Classification
|
Incompatibility is the most useful criterion. It refers to the ability of two plasmids to coexist
stably in the same bacterial cell. If they can, they are compatible. If they cannot, they are incompatible. Closely related plasmids are incompatible and fall into the same incompatibility group. The closely related plasmids are probably incompatible because they compete for the same replication machinery; compatible plasmids use different machinery from each other. This is useful for epidemiology |
|
|
A clinical isolate, bacterium A, resistant to streptomycin, erythromycin and
tetracycline, is mixed with bacterium B, which is known to be a mutant (because of chromosome mutation) resistant to nalidixic acid. From the mixture, you isolate bacteria resistant to all four antibiotics. How could you conclude that A was carrying a conjugative multiple-drug-resistant plasmid? What other experiment(s) could help you establish this? |
To begin with, why couldn't B be transferring Nal resistance to A? You would have to have some additional identifying characteristic (marker) to distinguish A from B.
For example, B could be antigenically distinct, and all the recombinants be of B antigenic type. Now, couldn't the multiple drug resistance result from a mutation in B? No, because mutations are rare, say one out of a million; therefore, a triple mutation would occur once out of 1018. Transfer of multiple drug resistance is typical of conjugative R plasmids. You could determine the plasmid profile of A, B and the B recombinants by gel electrophoresis to show that B recombinants have the same plasmid as A. Further, B recombinants could be used to transfer multiple drug resistance to another recipient. The transfer would be resistant to DNase (therefore not transformation) and require cell-cell contact between donor and recipient (therefore not transduction; diagnostic of conjugation). |
|
|
Transposons
|
AKA- Transposable Elements, Mobile Elements, "Jumping Genes"
Transposons are defined pieces of DNA that can move (transpose) as discrete units from one site in a DNA replicon to another site in the same replicon or in another replicon. The replicon can be the bacterial chromosome, or bacteriophage DNA, or a plasmid. Most transposons are never found free in nature; they are always inserted in some other DNA. Transposons are not replicons. Many carry drug-resistance determinants (translocatable drug- resistance elements) The smallest enterobacterial transposons, called insertion sequences or IS elements, code for their own transposition (i.e., for an enzyme, the transposase) but have no readily observable phenotype of their own (and thus are often detected when they insert into and inactivate another gene). Characteristics of Transposons: 1) They transpose (translocate, move) from one piece of DNA to another. Many can cross species barriers. 2) Transposons code for an enzyme transposase that mediates integration into the target DNA. (site specific, dot require other enzymes) 3) Transposition does not require homology between the transposon and the target site. 4) Insertion of the transposon into the target DNA is usually precise, i.e., there usually is no loss of target DNA (in fact, there is a duplication of a few base pairs of target DNA, either side of the inserted transposon, the precise number of bases being characteristic of the transposon). 5) Once inserted, the transposon is covalently joined to the target as part of the DNA double helix. 6) The ends of most transposons are necessarily inverted repeats or palindromes (allows transposase recognition. 7) When transposons integrate into a gene, they almost always inactivate that gene. 8) The transposons have mechanisms to repress transposition (and the introduction of a second copy of the same transposon). 9) Most transposons are never found free in nature; they are always inserted in some other DNA. Transposons are not replicons. |
|
|
Methods of Transposition
|
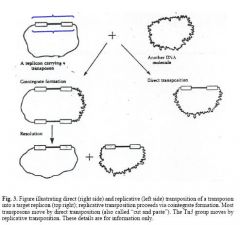
Replicative vs. Direct
|
|
|
Types of Transposons
|
1. IS elements
2. Composite/Compound Transposons - 2 IS elements have taken flanking positions with a functional gene in between them. They transpose as a unit (although each IS can rarely transpose by itself) and have a phenotype (note that only one IS has to functionally code for transposase). This is the mechanism for passing Antibiotic resistance. 3. Replicative Transposon- a transposon that requires a resolvase enzyme in addition to transposase. 4. Lysogenic Phage Mu- Lysogenized E. COli by inserting its DNA into many places on the chromosome. The DNA is very large for a tranposon, but is still not free, being flanked by small peices of host bacterial DNA. Mu = Mutation which it commonly causes. 5. Plasmid Free conjugative transposon- It induces bacteria to conjugate without any plasmid. The mechanism is not understood, but is different than the regular pheromone based plasmid system. Conjugation can even be induced between different species. They are widewpread and of clinical importance. |
|
|
Non Transposon Mobile Elements
|
Pathogenicity Islands (PAI's)- Large units that move as compact, distinct genetic packages. Often flanked by direct repeats, contain some kind of mobility gene. Typically they are present in pathogenic strains of a species, and absend in benign strains.
MRSA Mobile Elements (AKA SCCmec [Staphylococcal Cassette Chromosome with mec])- The methicillin resistance coded for by mecA represents a new kind of mobile element. There are several classes that are all big, and many contain IS elemetns/transposons and integrated plasmids. |
|
|
Site Specific Recombination
|
Transposition is one form of site specific recombination, which does not require homology and in E. Coli does not require RecA. Lysogeny and Phase Variation are also considered sire specific recombinatoin
|
|
|
An IS element inserts into the promoter region of the lactose operon.
a. What would probably happen to transcription of the structural genes? b. If the IS element precisely excises, what would happen to transcription of the structural genes? |
a, transcription would be prevented; b, transcription would be restored. Indeed, it was a similar observation with the galactose operon which led to the discovery of IS elements in bacteria. The galactose-negative phenotype was at first interpreted to be due to a deletion mutation, because many products of the galactose operon were missing, suggesting a deletion. But the reappearance of a complete parental phenotype could not be due to back mutation of deletions, because back mutations cannot restore the exact sequence of DNA. Note that transposons
can excise precisely. |
|
|
Which of the following is true?
Insertion sequences by themselves: a. carry drug R genes b. are part of composite transposons c. are replicons |
b. are part of composite transposons
|
|
|
Which of the following is true?
An R plasmid can: a. code for antibiotic-destroying enzymes b. change cell permeability to antibiotics c. be a sex factor d. be transferred by transduction |
All of the Above
a. code for antibiotic-destroying enzymes b. change cell permeability to antibiotics c. be a sex factor d. be transferred by transduction |
|
|
Which of the following is true?
A transposon: a. can transfer drug resistance b. can move from plasmid to chromosome c. is covalently linked as linear double-stranded DNA within its target d. is a replicon |
a, b, and c
a. can transfer drug resistance b. can move from plasmid to chromosome c. is covalently linked as linear double-stranded DNA within its target |
|
|
No. What is depicted is the normal or wild genotype (by convention denoted by
the superscript "+") of E. coli. In this normal state E. coli is 1) inducible for transcription of the lactose operon, 2) inducible for production of the three enzymes, and 3) inducible for lactose fermentation and utilization. |
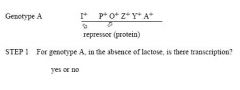
Image
|
|
|
No. What is depicted is a mutation in a regulator gene of the lactose operon. In
this state E. coli is 1) constitutive for lactose fermentation and utilization, the reason being that, 2) the mutation is in the repressor gene which results in a bad repressor. 3) Another way of stating the situation is to say that the operon is derepressed. |

For B, is lactose needed for transcription of the operon?
|
|
|
No. What is depicted is a mutation in the operator of the lactose operon. In this
state E. coli is 1) constitutive for lactose fermentation and utilization, the reason being that, 2) the mutation is in the operator (Oc for operator constitutive), in which case the Z, Y and A genes are continuously transcribed (they are "on"). 3) This is another case of a derepressed operon. |

Image
|
|
|
Yes, even the non-functional proteins (enzymes) of the Z- and Y- genes are made.
Under special experimental conditions, non-functional proteins can be detected in E. coli cells as antigenic equivalents of functioning proteins. [Exceptions are polar mutations in a gene, which affect transcription of the downstream gene(s); these are beyond the scope of the course.] Let's analyze. There is a double dose of repressor, but that's okay. When you add lactose to a culture of E. coli, there are always more than enough lactose molecules to tie up the repressor. The P and O regulator genes are okay, normal. There is a single dose of β-galactosidase and permease, but that's okay - enough for the cell to be lactose positive. There's an extra dose of transacetylase (whose role in utilization of lactose is unknown), and that's okay. The answer is yes. |

For D, in the presence of lactose, are the structural genes transcribed? yes or no
For D, in the presence of lactose, is the E. coli lactose positive, i.e., does it ferment and utilize lactose? (note, there are 2 mutations) yes or no |
|
|
No, because a repressor can act in the trans position (N.B., this is NOT the same
as TRANSPOSITION [one word] of a transposon), or across from the gene which codes for it. In this case, the repressor proteins act on the O regions of the chromosome (C) and of the F genote. That is, I+ is dominant in trans over I-, or, put another way I- is recessive to I+. (Genote is a convenient genetic term for a set of genes) |

For E, in the absence of lactose, are the structural genes transcribed? yes or no
|
|
|
Let's analyze. The Z, Y and A genes of the F genote could be transcribed into
mRNA for non-functioning proteins. So, the answer is yes. |

For F, in the absence of lactose, are any structural genes transcribed? Look
carefully at the F genote - now answer yes or no. |
|
|
An operator can only act as a switch for adjacent genes, i.e., when it is in cis
position to those genes. Therefore, the Oc genotype of the F genote cannot turn on the Z+, Y+, A+ genes of the chromosome. O+ is cis dominant. Similarly, Oc is cis dominant. lacO is a cis element LacI is a trans-acting factor |

For F, in the absence of lactose, are functional proteins made? The answer is no, but do you know the reason?
|
|
|
a) Yes, because the product of the C+ gene, the apo-activator, is present. The Cmutation is recessive (i.e., C+ is dominant in trans over C-). b) No, L-arabinose is needed.
|
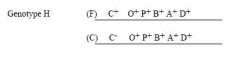
For H, is there transcription:
a) in the presence of arabinose; b) in the absence of arabinose? |
|
|
a) Yes, for the same reason given for 13a.
b) Yes, because the apo-activator works in the trans position. Note: the B-, A-, and D- genes of the F genote may produce nonfunctioning, B-, A-, D-, protein products. c) No, L-arabinose is needed. d) No, L-arabinose is needed. |

For I, in the presence of arabinose:
a) is there transcription; b) are functional proteins produced? For I, in the absence of arabinose: c) is there transcription; d) are non-functional proteins produced? |
|
|
In haploid E. coli, how does the result of a deletion mutation of the lacI gene
differ from that of a deletion mutation of the araC gene? |
Deletion mutations of lacI results in no production of a repressor. Therefore,
expression of the lactose operon is constitutive, i.e., the structural genes are transcribed even in the absence of lactose. Deletion mutations of araC result in no production of the activator. Therefore the arabinose operon is off, i.e., the structural genes are not transcribed even in the presence of arabinose. In other words, the opposite result to that when lacI was deleted; this is a general difference between control genes exerting positive and negative control. |
|
|
Phase Variation
|
A genetic alteration (not mutation?) which confers a phenotypic advantage over another genetic conformation, such as evasion of host immune system. There are 3 main mechanisms - Inversion, Cassette Like, and Strand Slippage
|
|
|
Phase Variation - Inversion
|
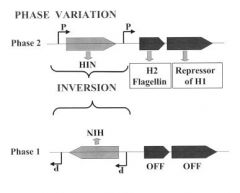
The classic example of phase variation is the switch in flagellar antigens (the H antigens) of Salmonella. In it, a small piece of DNA (about 1 kbp) inverts, changing the bacterium from one phase to another. In Salmonella in phase 2 a promoter within the invertible element drives the
transcription of the genes for H2 flagellin and for the repressor of the gene for H1 flagellin (which is located elsewhere on the chromosome). In phase 1 the promoter points in the other direction so that neither the H2 flagellin nor the repressor are made. This change permits transcription of the gene for the H1 flagellin. The inversion (phase variation) is catalyzed by the product of the hin gene, which is sometimes called the Hin recombinase. The phase variation is not a form of transposition, although the Hin recombinase has extensive similarity to transposases. A similar system to the one described above is found in the control of type 1 fimbriae of uropathogenic E. coli (UPEC). In Bacteroides fragilis, 13 different regions invert, all controlling surface structures, and all controlled by one recombinase. |
|
|
Cassette Like- Phase Variation
|
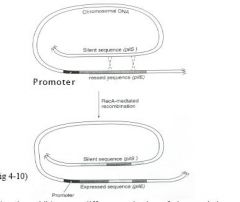
Recombinatorial mechanisms operate in Haemophilus
influenzae, Neisseria gonorrhoeae and Neisseria meningitidis. For example, N. gonorrhoeae uses a cassette-like mechanism to vary phase with respect to pilus formation. Scattered around its chromosome are multiple non-expressed genes (they have no promoter) for pilus formation (pilS genes, where S means silent); some of these are truncated or otherwise mutated. Typically, one copy, pilE, is expressed (E for expressed) because it is adjacent to a promoter. Occasionally, the pilE gene and a pilS gene recombine so that a slightly different pilus protein, or a truncated protein or no protein, is made. This mechanism enables N. gonorrhoeae to evade the host immune response. Non-piliated strains are non-pathogenic. |
|
|
Strand Slippage - Phase Variation
|
Haemophilus influenzae, Neisseria gonorrhoeae and Neisseria meningitidis also exhibit a very different mechanism of phase variation - strand slippage during DNA replication, transcription or translation - as do other species such as Bordetella
pertussis, Helicobacter pylori and Vibrio cholerae. Phase variation of N. gonorrhoeae opa genes gives an example of strand slippage during DNA replication. The Opa proteins are a set of outer membrane, adherence proteins. They get their name because of the opacity of colonies as a result of the adherence of the bacteria. There are about 12 related opa genes. Each has 7 to 28 repeats of the penta-nucleotide sequence CTCTT in the 5’ end of the gene. The number of repeats can change as a result of “stuttering”, or slippage, during DNA replication. As a consequence, there is generally a change in reading frame. Only one reading frame gives full-length, functional protein. Different opa genes are turned off or on as a consequence of this stuttering. Given about 12 opa genes (the number varies strain to strain), there can be a lot of variation in the Opa proteins being expressed. This mechanism enables N. gonorrhoeae to evade the host immune response. Opa proteins are important for virulence, though their precise role is not established. |
|
|
Bacteriophages
|
Bacteriophage (phage) are bacterial viruses. There are different morphological types of phages. The capsid or outer coat may be polyhedral,
round or filamentous. There may be tail structures. They are very small and cannot be seen by light microscopy. The outer structures are protein and the inner structures are nucleic acid - usually double-stranded DNA, but some phage have single-stranded DNA, and others RNA. Ex. Escherichia coli phage T2, T4 and λ all have double-stranded DNA; M13 has single-stranded DNA and MS2 has single-stranded RNA. Phage nucleic acids code for anywhere between 3 and 200 proteins. |
|
|
Lytic/Lysogenic Cycle
|
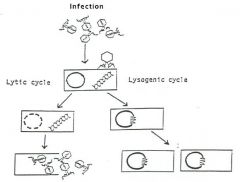
Phages invade bacteria and enter either the lytic or lysogenic phase.
|
|
|
Lytic Bacteriophage Infection
|
1. Phage adsorbs to bacteria (by tail fibers if present). Adsorption depends upon specific receptor sites. Presence of the receptor sites accounts for host range (Phage commonly have a very narrow strain/species range).
2. The phage particle acts like a syringe, injecting its nucleic acid into the host bacterium. The nucleic acid gets inside the bacterium, but the capsid and other structures, if any, remain outside. 3. Double-stranded DNA (most phages) often replicates bidirectionally, in a semi-conservative manner. Phage with single-stranded nucleic acid (DNA or RNA) first makes a complementary copy, a replicative form (RF), from which the ssDNA and ssRNA are made (using the rolling circle) 4. Phage nucleic acid codes for phage structural proteins, including the proteins of the capsid (or head) and any additional structures - tails, baseplates, tail fibers, etc. All of these are assembled separately, and note that the head is formed before instilation of the genetic material. 5. Phage Head Packaging - Different phage have different mechanisms. The objective is to have one genome per phage particle. Some phage package a "headfull" of DNA Ex. Phage lambda will package 40 to 50 kbp DNA provided it is flanked by cos sites). 6. Bursting - The vegetative phage burst out (with the exception of some filamentous phage which slowly release through the cell membrane just like influenza virus). The burst size is the number of phage particles produced from a single infecting phage particle after a single round of infection, and is characteristic of the particular phage. |
|
|
Roling Circle Replication
|
Used by both single stranded and some double stranded phages; this is not the conventional DNA replication, in that both strands are not copied. Plus strands are being made and peeling off a circular Replicative Form (RF).
|
|
|
Phage Assay
|
In principle: Mix a dilution of a phage with a suspension of a susceptible host bacterium. Allow the phage to absorb to the bacteria for about 5 minutes. The mixture is then usually added to
melted nutrient soft agar and poured. After solidification incubate and the bacteria grow throughout the agar, and the phage grows in each infected bacterium. The phage burst out and infect surrounding bacteria and cycles of burst/infectiont. Plaques, each representing infection started by one phage particle, appear in the lawn of bacterial growth as a clear area, each plaque contains millions of phage particles, and are analogous to colonies of bacteria. Often phage titres are given as PFU (Plaque Forming Units). Phages can be classfied as virulent of Temperate |
|
|
Virulent Phages
|
Virulent phages only go through a lytic cycle of infection. Virulent phage give clear plaques.
|
|
|
Temperate Phages
|
Temperate (or lysogenic) phages have an option – either a lytic cycle or a lysogenic cycle.Temperate phage typically give turbid plaques reflecting a balance
between the lytic cycle and lysogeny. The turbidity is caused by lysogenized bacteria growing in the plaques. (Lysogenized bacteria are immune to superinfection by the same phage.) Lambda of E. Coli is the prototype. It has sticky ends (cos sites) that mark the end of the phage. Establishment of lysogeny involves a site-specific recombination between the attP site of the phage DNA circle and the attB site of the host chromosome. The result is that the phage DNA becomes part of the chromosome, and is replicated as such. A complex interplay between phage repressors and activators and their target cis elements determines whether the lysogenic or lytic path is followed. (Host factors and environment are also involved, but do not worry about the details). DNA damage can induce, or prophage spontaneously can initiate, a lytic cycle. |
|
|
Lysogeny
|
That state in which a temperate (or lysogenic) phage is maintained in a
quiescent state, the prophage; in it, the phage DNA replicates with the host chromosome. |
|
|
Prophage
|
the name for phage DNA in the lysogenic state within the bacterial host. In the case of
lambda (see below) the prophage DNA becomes part of the chromosome of the lysogenized bacterium. For some other phage, the prophage DNA is a plasmid that replicates separately from the chromosome of the lysogenized bacterium. |
|
|
Immunity to Superinfection
|
A consequence of lysogeny is immunity to superinfection by the same phage because of a
protein repressor encoded by the prophage. The λ repressor, protein CI, represses the expression of all λ prophage genes - except the gene for CI. The CI repressor also represses the genes of any superinfecting phage λ; note that it will not repress the genes of other types of phage. Immunity is different from the resistance of mutant bacteria, in which the phage receptor site has been lost or altered by mutation. |
|
|
Generalized Vs. Specialized Transduction
|
Generalized - during
replication of phage, the host chromosome breaks into fragments. Occasionally, by mistake, these fragments are packaged into the phage capsid instead of phage DNA. Any bacterial gene (or several closely linked genes) can be transduced from one host to another, by the resulting particles which contain no phage DNA. Specialized - only a select few genes (or just one gene) are transduced by the phage; they are those genes close to the site of prophage insertion in the chromosome. Because the acquisition of the segment of bacterial DNA occurred at the expense of a portion of phage DNA, these particles may not be able to complete a normal infection process and are termed "defective". A defective phage can only complete and infection/form plaques if it infects a bacterium along with a normal or "helper" phage (complementation). Initially this happens at a low freq, however after being formed and selected high frequency transducing (HFT) lysates can be prepared. Note also that Specialized happens at a higher frequency than general. |
|
|
You pick the center of a turbid plaque of phage X, streak a nutrient agar plate, and subsequently get colonies of bacteria which survived phage X in the plaque. Using one
colony as an example, how would you determine whether the survivor is lysogenic for phage X or resistant to phage X? What about growing a culture of the survivor and spotting phage X on a lawn of the survivor? |
Spotting phage X on a lawn of the survivor would probably tell you nothing. Phage
would not lyse a survivor that is lysogenized and therefore immune, or one that is resistant. Take a broth culture of the survivor, preferably get rid of the bacteria (by filtration), and spot the broth on a lawn of the original susceptible host. The broth of a culture of a lysogenic survivor would have some free phage (spontaneous lysis), and the phage would produce plaques on the susceptible host, whereas a phage-resistant mutant is not lysogenic and would not shed phage into broth. An added piece of evidence would involve showing that an immune (lysogenic) survivor can nevertheless adsorb phage X (although this superinfection is not productive), whereas a resistant mutant survivor can't even adsorb phage. How would you run an adsorption experiment? Think about it. |
|
|
Which is better suited to map the chromosome of E. coli K12 - generalized or
specialized transduction? |
Generalized transduction, because any set of closely linked genes can be transferred. This does result in mapping by scoring for adjacent (linked) genes. For example, if A and B are co-transduced, and B and C are co-transduced but A and C are not co-transduced then the map is A-B-C (not
A-C-B). |
|
|
Phage Conversion
|
With some phage lysogeny alters metabolism of the host, or changes host surface antigens. This phenomenon is called lysogenic (or phage) conversion, and can be of great importance to
pathogenicity. The classic example is the β phage of Corynebacterium diphtheriae. C. diphtheriae is non-pathogenic unless lysogenized by phage β (exotoxin-diptheria). Vibrio cholerae appears to have been derived from non-pathogenic V. cholerae through two successive conversions, one providing a pilus, and the other a toxin. Other diseases resulting from conversin include Group A strep, salmonells, EHEC, and Botulism. |
|
|
Phage Typing
|
This is a scheme for typing strains of, for example, Staphylococcus aureus and Salmonella typhi based on the pattern of susceptibility (or resistance) to a battery of phages. The susceptibility
pattern is affected by lysogenic immunity, altered receptor sites, and phages with various host ranges. Phage typing can be used in epidemiology to track the source of an infection (the index case). Phage typing is not widely used clinically today, but may be it the future (if we can get it to work) |
|
|
There are so many similarities between phage and plasmids - rolling-circle DNA replication, ability to insert into chromosome, toxigenic and antigenic conversion, etc. (you
can continue the comparison) - that it can be speculated that a. phage evolved from plasmids which gained the ability (by mutation or recombination) to make capsids, tails, etc. b. plasmids are mutants of phage which lost the ability to make capsids, tails, etc. |
No one knows...Chicken and the Egg.
|

