![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
148 Cards in this Set
- Front
- Back
|
Microbial Molecular Biology: Why should you care? |
If we understand the differences between eukaryotic and prokaryotic molecular biology we can exploit these differences to our benefit (for drugs, vaccines, biotech, etc.) |
|
|
How can we use the differences between eukaryotic and prokarotic molecular biology for our benefit? |
drugs, vaccines, biotech, etc. |
|
|
What is the central dogma of biology |
DNA -> RNA -> Protein Replication -> Transcriptions -> Translation |
|
|
Define Dogma |
A principle or set of principles laid down by an authority as incontrovertibly true (Oxford American Dictionary) |
|
|
What violated the central dogma of biology? |
The concept of reverse transcription, where RNA is used as a template to make DNA, violated the dogma. It took a white to catch on. |
|
|
What are the four nucleotide bases of DNA? |
Adenine (A) Guanine (G) Cytosine (C) Thymine (T) |
|
|
What kind of backbone does DNA have? |
Deoxyribose sugar + phosphate backbone |
|
|
How are the strands of DNA held together? |
Strands held together by hydrogen bonds between A+T (2 bonds) and G+C (3 bonds) |
|
|
How many bonds between Adenine and Thymine? |
2 |
|
|
How many bonds between Guanine and Cytosine? |
3 |
|
|
Who was the first person to create an X-ray diffraction image of DNA |
Rosalind Franklin |
|
|
Who published the double-helix model for DNA and what year? |
James Watson and Francis Crick published their double-helix model of DNA structure in 1954 (Franklin's data was essential) |
|
|
What is the shape of DNA? |
DNA strands wind around each other to form a double helix |
|
|
What are the two distinct groves that DNA forms? |
Forms 2 distinct grooves - a major groove and a minor groove |
|
|
Where do proteins usually interact on a DNA structure? |
Proteins (usually) interact with the major groove |
|
|
DNA in a living organism typically exists in a "_____/_____" helix |
right/handed |
|
|
What does a right handed DNA look like? |
Most DNA molecules, like most screws, are right-handed |
|
|
The DNA double helix is _____ |
Twisted |
|
|
About how many bp per twist in DNA? |
~10 bp/turn |
|
|
What does introducing or removing additional DNA twists lead to? |
Supercoiling |
|
|
Define supercoiling |
Introducing or removing additional twists that imparts strain on the DNA molecule |
|
|
Define positive supercoiling |
Extra twists (overwinding) |
|
|
Define negative supercoiling |
Subtraction of twists (underwinding) |
|
|
Most DNA in living organisms is ______ supercoiled. |
Negatively |
|
|
Supercoiling allows lots of DNA to be packed into a bacterial cell (T/F) |
True |
|
|
The bacterial chromosome (nucleoid) contains multiple loops held by _____/_____ |
anchoring/proteins |
|
|
What does each loop of chromosome contain? |
Each loop contains supercoiled DNA |
|
|
How much linear DNA does an E. Coli nucleoid contain? |
The E. Coli nucleoid contains more than 1mm of linear DNA - at least 400 times longer than the cell itself |
|
|
If E. Coli was the size of Andre the Giant, how long would its DNA be? |
Its DNA would be more than half a mile long! |
|
|
DNA is wrapped around proteins called _____ to form a nucleosome. |
histones |
|
|
How much DNA is wrapped around a histone? |
Wraps around 200 bp of DNA |
|
|
Nucleosomes _____, with higher order folding of DNA, to form _____ |
aggregate chromatin |
|
|
Supercoiling mediated by DNA gyrase (picture) |
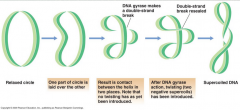
|
|
|
In prokaryotes, _____/_____ ( a class II topoisomerase) introduces _____ supercoils into the DNA |
DNA/Gyrase negative |
|
|
What class is DNA Gyrase? |
A class II topoisomerase |
|
|
What happens when a DNA molecule is exposed to DNA Gyrase? |
The DNA molecule is broken and resealed on the opposite side of the strand |
|
|
DNA Gyrase is found in both prokaryotes and eukaryotes. (T/F) |
False. DNA Gyrase found only in prokaryotes |
|
|
What is the net effect of the mechanism of type II topoisomerases? |
Net effect is insertion of one negative supercoil |
|
|
What makes a good target for antibiotics? |
1. Inhibition of cell wall synthesis 2. Inhibition of protein synthesis 3. Inhibition of nucleic acid replication and transcription 4. Injury to plasma membrane 5. Inhibition of synthesis of essential metabolites |
|
|
Name 4 antibiotics that inhibit cell wall synthesis |
penicillin cephalosporin bacitracin vancomycin |
|
|
Name 4 antibiotics that inhibit protein synthesis |
Chloramphenicol erythromycin tetracyclines streptomycin |
|
|
Name 2 antibiotics that inhibit nucleic acid replication and transcription |
quinolones rifampin |
|
|
name 1 antibiotic that hurts the plasma membrane |
polymyxin B |
|
|
Name 2 antibiotics that inhibit the synthesis of essential metabolites |
sulfanilamide trimethoprim |
|
|
name 3 antibiotics that target DNA Gyrase |
Quinolones Fluoroquinones Aminocoumarins |
|
|
Why is DNA Gyrase a good antibiotic target? |
Not present in most eukaryotic cells = good target |
|
|
Some prokaryotes have reverse gyrase (T/F) |
True |
|
|
Define reverse gyrase |
is a topoisomerase that introduces positive supercoils |
|
|
Where is reverse gyrase mostly found? |
Mostly found in Archaea that grow at extremely high temperatures |
|
|
Why would Archaea that live in high temperatures want reverse gyrase? |
May protect the DNA from heat denaturation - positive supercoiled DNA is harder to denature |
|
|
What are the characteristics of a prokaryotic chromosome? |
Circular (usually) Usually only one Do not have telomeres Do not have multiple copies of genes - usually Greater than 90% of the genome consists of genes encoding for known proteins - very little repetitive sequence |
|
|
What shape is the prokaryotic chromosome (usually)? |
Circular |
|
|
Do prokaryotics have more than one chromosome? |
Usually only one |
|
|
Do Prokaryotic chromosomes have telomeres? |
no |
|
|
How much of a prokaryotic genome consists of genes that encode for known proteins? |
90% |
|
|
What are the characteristics of a Eukaryotic chromosomes? |
Linear chromosomes More than one per cell (23 pairs in humans) Telomeres on the ends required for complete chromosome replication Often contain multiple copies of each gene Contain more DNA than necessary, not all function (or known to be) <3% of the human genome encodes protein |
|
|
What shape are eukaryotic chromosomes? |
Linear |
|
|
Why do eukaryotic chromosomes have telomeres? |
Telomeres on the ends required for complete chromosome replication |
|
|
About how much of the human genome encodes protein? |
<3% |
|
|
How many bp in Bacterial genomes? |
0.6-9.4 million bp |
|
|
How many bp in human genome? |
3 billion bp |
|
|
How much bigger is the human genome vs E. Coli's genome? |
1000X as large as E. Coli |
|
|
How many more genes does a human genome have vs. E. Coli |
~6x genes: ~23,000 (human) vs. 4,000 (E. Coli) |
|
|
E. Coli genes use less DNA sequence. (T/F) |
True |
|
|
How many bp go into a typical bacterial gene? |
About 1000 bases |
|
|
Define plasmids |
Plasmids are small circular extrachromosomal units of DNA in bacteria that often encode genes for antibiotic resistance |
|
|
How are plasmids exchanged? |
Plasmids may be exchanged between bacteria of the same or different species allowing antibiotic resistance to spread rapidly |
|
|
Prokaryotes have Mitochrondria and Chloroplasts. (T/F) |
False. Eukaryotes have mitochondria and chloroplasts |
|
|
Mitochondria and Chloroplasts contain their own _____ material. |
Genetic |
|
|
Mitochondria and Chloroplasts replicate _____ of the chromosomes |
Independently |
|
|
Mitochondria and Chloroplasts have everything necessary for protein synthesis. (T/F) |
True |
|
|
What do mitochondria/chloroplasts have in order to make them capable for protein synthesis? |
ribosomes, tRNA, etc. |
|
|
Why are Mitochondria/chloroplasts not independent of the cell? |
most of their proteins are coded by the chromosomes in the nucleus |
|
|
Mitochondria and chloroplasts are independent of the cell (T/F) |
False. NOT independent of the cell - most of their proteins are coded by the chromosomes in the nucleus |
|
|
Semiconservative replication (picture) |
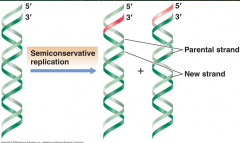
|
|
|
DNA replication (picture) |

|
|
|
All ______/______ synthesize in the _____ to _____ direction |
DNA/Polymerases 5' 3' |
|
|
No known DNA polymerase can initiate DNA synthesis. (T/F) |
True |
|
|
______ adds an RNA primer to the _____ end |
Primase 5' |
|
|
In what direction does DNA polymerization occur? |
5'->3' |
|
|
Only ______ strand is oriented ______ for polymerization to proceed ______. |
strand correctly 5'->3' |
|
|
Define leading strand |
The one strand that is oriented correctly for polymerization to proceed 5' -> 3' In this strand DNA polymerization is continuous |
|
|
Define the lagging strand |
DNA polymerization is discontinuous because the strand is not oriented correctly |
|
|
DNA Replication Fork (picture) |
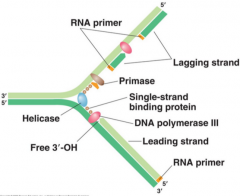
|
|
|
Does does primase do? |
Primase adds an RNA primer to the lagging strand at multiple points along the strand as it is unwound |
|
|
______/_____ on the lagging strand are later joined together |
Okazaki/fragments |
|
|
Sealing the fragments on the lagging strand (picture) |
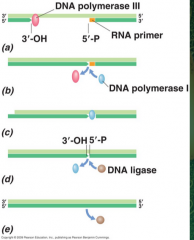
|
|
|
How are the fragments of the lagging strand sealed? |
DNA polymerase III stops when it reaches a downstream primer DNA polymerase I has 5' to 3' nuclease activity. It removes the RNA primer and adds DNA DNA ligase makes the final phosphodiester bond |
|
|
Replication of Circular DNA (picture) |
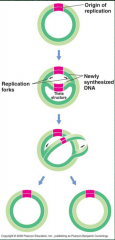
|
|
|
Origin of replication |
A specific sequence of approx. 300 bp seen in circular replication |
|
|
Replication is bidirectional (T/F) |
True |
|
|
What forms during replication of circular DNA |
Forms theta structures in circular DNA |
|
|
Explain the replisome |
coordinated synthesis of both strands at once Contains two DNA polymerase III and other molecules needed for replication Loops the lagging strand Replisome is anchored near the midpoint of the cell Note that the DNA moves, not the replisome! |
|
|
Explain whole-genome sequencing |
Break genome into thousands of pieces that are easy to sequence Determine sequence of many short pieces at random Computer determines sequence overlap to recreate entire genome sequence |
|
|
Whole-genome sequencing (picture) |
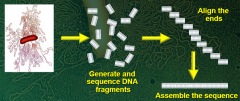
|
|
|
How many polymerases do Eukarya have? |
Eukarya have 3 RNA polymerases |
|
|
How many polymerases do Bacteria and Archaea have? |
Bacteria and Archaea have one RNA polymerase |
|
|
What are the two forms of Bacterial RNA? |
core enzyme Holoenzyme |
|
|
What is the role of the sigma subunit? |
Role of sigma subunit is to recognize the promoter site on the DNA for initiation of RNA synthesis |
|
|
sigma subunit is tightly bound to the other subunits (T/F) |
False. Sigma subunit is not tightly bound to the other subunits |
|
|
What does sigma does? |
Sigma locates the initiation site, then dissociates |
|
|
What do core enzymes do? |
Core enzyme synthesizes the RNA |
|
|
Define promoter |
Site recognized by RNA polymerase |
|
|
DNA is unwound and RNA is synthesized from one of the strands. (T/F) |
True |
|
|
Several different sigma factors - recognize distinct promoters (T/F) |
True |
|
|
In a single promoter there are ______ sets of ______ that are conserved. |
2 sequences |
|
|
What are the 2 set of sequences that are conserved in a single promoter? |
The pribnow box (at -10) - TATAAT -35 sequence - TTGACA |
|
|
Different sigma factors are used in response to different ______ conditions |
environmental |
|
|
Name and define E. Coli haas 7 sigma factors |
70 - "housekeeping" sigma factor, transcribes most genes in growing cells 38 - starvation/stationary phase genes 28 - genes involved in flagellar synthesis 32 - genes involved in heat 24 - genes associated with extracytoplasmic stress 54 - Nitrogen-limitation response genes 19 - genes mediating iron transport |
|
|
Different sigma factors can act as '_____/____' |
virulence/factors |
|
|
Define unit of transcription |
The section of DNA that is transcribed into RNA |
|
|
Unite of transcription (picture) |
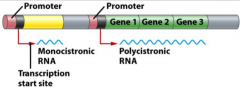
|
|
|
Can a unit of transcription contain more than one gene? |
Yes |
|
|
Define polycistronic RNA |
When there is more than one gene on a unit of transcription Common in prokaryotes |
|
|
Define operon |
A group of related genes transcribed as a polycistronic RNA |
|
|
What does Rifamycins do? |
inhibit bacterial RNA polymerase |
|
|
Define RNA translation |
Process by which the RNA sequence is converted into protein sequence |
|
|
What does RNA translation use to determine which amino acid comes next? |
Use the genetic code to determine which amino acid comes next |
|
|
Each set of _____/_____ specifies an amino acid |
3/nucleotides |
|
|
3 nucleotides are called a ______ |
Codon |
|
|
Define codon |
set of 3 nucleotides that specify an amino acid |
|
|
How many possible 3 letter codons are there? |
64 (4^3) |
|
|
Define degeneracy |
more than one codon codes for an amino acid |
|
|
Define Wobble |
the 3rd nucleotide can vary |
|
|
Most organisms use what code? |
The "universal code" |
|
|
Some organisms and organelles use slight variations of the code. (T/F) |
True |
|
|
Animal mitochondria use the stop codon _____ to encode _____ |
UGA tryptophan |
|
|
What does animal mitochondria using the stop codon UGA to encode tryptophan an indication of? |
Indication of different evolutionary history |
|
|
In Mycroplasm (_____) and Paramecium (_____) "_____" (stop) codons can encode _____/_____ |
bacteria eukarya nonsense amino/acids |
|
|
In certain cases ______ codons can encode rare _____/_____ such as _____ and _____ |
nonsense amino/acids selenocysteine pyrrolysine |
|
|
_____ reads the base pairs on the codon |
tRNA |
|
|
How many nucleotides on transfer RNA? |
73-93 nucleotides |
|
|
Describe tRNA |
73-93 nucleotides, chemically modified bases, extensive secondary structure |
|
|
Protein synthesis is mediated by what? |
Mediated by the ribosome, a ribonucleoprotein complex consiting of ribsomal RNAs and proteins |
|
|
What are the three steps of protein synthesis? |
1. Initiation 2. Elongation 3. Termination |
|
|
Ribosomes of mitochondria and chloroplasts of eukaryotes are similar to prokaryotic ribosomes (T/F) |
True |
|
|
How is the initiation of translation started? |
mRNA, tRNA, plus initiation factors assemble on the 30S subunit Associates with 50S ribosomal subunit |
|
|
How is the correct reading frame determined for prokaryotes? |
the 16S rRNA binds to the Shine-Dalgarno sequence, a sequence of 3 to 9 nucleotides just proceding the initiation codon in the mRNA
|
|
|
How is the correct reading frame determined in Eukaryotes? |
Ribosome binds to the 5' cap on the mRNA |
|
|
Define Intiation |
First charged tRNA binds in P-site (Peptide site) |
|
|
Define elongation |
A second charged tRNA binds in the A-site (acceptor) Peptide bond formation occurs between amino acids in P- and A-sites |
|
|
Define translocation |
Uncharged tRNA in P-site moved to E-site (exit site) and tRNA in A-site moves to P-site Uncharged tRNA released from E-site and next charged tRNA binds to A-site |
|
|
When does termination of protein synthesis occur? |
Occurs when the ribosome reaches a stop codon No t RNA binds |
|
|
______/_____ recognize the stop codon and cleave the attached polypeptide |
Release/factors |
|
|
During the termination of protein synthesis what does the ribosome dissociate into? |
Ribosome dissociated into 30S and 50S subunits, which can then form new initiation complexes |
|
|
Define the polyribosome |
Also known as the polysome Several ribosomes can translate a single mRNA simultaneously This speeds up the process and generates additional protein product |
|
|
The polyribosome was first described by whom and what year? |
The polyribosome was first described by Warner, Knopf & Rich in 1963 |
|
|
Why do many antibiotics target the bacterial ribosome? |
Rely on differences between prokaryotic and eukaryotic ribosomes |

