![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
127 Cards in this Set
- Front
- Back
|
Gene
|
Series of DNA nucleotides that generally codes for production of a single polypeptide or mRNA, rRNA or tRNA
|
|
|
Eukaryote genes vs. prokaryote genes
|
Eukaryotes: have more than one copy of some genes
Prokaryotes: have only one copy of each gene |
|
|
Genome
|
Entire DNA sequence of an organism
|
|
|
Purines
|
2 ring structures
Adenine and guanine Binds to pyrimidines |
|
|
Pyrimidines
|
single ring structures
Thymine or Uracil (RNA) and cytosine Binds to purines |
|
|
Phosphodiester bond
|
binds nucleotides together between the 3rd C of one deoxyribose and the 5th C of the other
Creates the sugar-phosphate backbone of a single DNA strand |
|
|
5' --> 3' Directionality, what is attached to each?
|
5' and 3' indicate the C #s on the sugar
3' C is attached to an OH group 5' C is attached to a phosphate group |
|
|
Length of DNA strand is measure in:
|
base pairs (bp)
|
|
|
Base pairing
|
H-bonds form between specific purines and pyrimidines
Adenine forms 2 H-bonds to thymine Guanine forms 3 H-bonds to cytosine |
|
|
Double helix
|
Formed by complementary strands of DNA
Major and minor grooves Each groove spirals once around double helix for every 10 bp |
|
|
What makes up a nucleotide?
|
1. phosphate group
2. 5-C sugar 3. nitrogenous base |
|
|
Semiconservative DNA replication
|
When new strand is created, it contains one strand from original DNA and one newly synthesized strand
|
|
|
Bidirectional DNA replication
|
2 replisomes proceed in opposite directions from an origin along the chromosome
|
|
|
Semidiscontinuous
|
Property of replication
One strand is continuous and other strand is fragmented |
|
|
DNA polymerase
|
Enzyme that builds the new DNA strand
Can only add nucleotides to an existing strand (needs primer) Creates new strand in the 5' --> 3' direction (downstream) Reads parental strand in the 3' --> 5' direction (upstream) |
|
|
RNA Primer
|
Created by RNA polymerase (primase)
Approximately 10 RNA long initiation strand to which DNA polymerase adds nucleotides to |
|
|
Lagging strand
|
Continuously interrupted strand
Restarted with a new primer Made from serious of disconnected strands (Okazaki fragments, which are 100-200 nucleotides long in eukaryotes, 1000-2000 in prokaryotes) |
|
|
Leading strand
|
Continuous new strand
|
|
|
DNA ligase
|
Moves along lagging strand and ties Okazaki fragments together to complete the polymer
|
|
|
5 steps of DNA replication:
|
1. Helicase unzips double helix
2. RNA polymerase builds a primer 3. DNA polymerase assembles leading and lagging strands 4. Primer are removed (exonuclease) 5. Okazaki fragments are joined by DNA ligase It is fast and accurate |
|
|
Telomeres
|
Repeated 6 nucleotide units on the ends of eukaryotic chromosomal DNA
From 100 to 1000 unites long Protect chromosomes from erosion by repeated replication |
|
|
RNA
|
Ribonucleic acid is identical to DNA structure except:
1. 2nd C on pentose is not deoxygenated (RNA has OH group attached) 2. Single stranded 3. Contains pyrimidine Uracil (U) instead of thymine 4. Not confined to nucleus, can move through nucleus pores |
|
|
rRNA
|
Combine with proteins to form ribosomes
Synthesized in nucleolus |
|
|
Ribosomes
|
Cellular complexes that direct synthesis of proteins
|
|
|
tRNA
|
Collects amino acids in cytosol
Transfers amino acids to ribosomes for incorporation into proteins |
|
|
Differences between DNA & RNA:
|
-DNA is only in nucleus and mitochondrial matrix and RNA is also in cytosol
-There is one major type of DNA and there are three major types of RNA -Replication requires a primer and transcription requires a promoter |
|
|
Promoter
|
sequence of DNA nucleotides that designates a beginning point for transcription for RNA polymerase
|
|
|
Transcription
|
Takes place in 2 places:
1. nucleus 2. mitochondrial matrix Only template strand (- or antisense strand) of DNA double helix is transcribed Coding strand (+ or sense strand) of DNA protects template strand from degradation |
|
|
Initiation
|
Beginning of transcription
Transcription initiation complex, which includes RNA polymerase, binds to to promoter on DNA strand |
|
|
Elongation
|
RNA polymerase transcribes template strand of DNA into complementary RNA strand
RNA polymerase reads 3' --> 5' RNA polymerase creates 5' --> 3' |
|
|
Termination
|
End of transcription required special termination sequence and special proteins to dissociate RNA polymerase from DNA
|
|
|
Activators
|
Proteins that bind to DNA close to promoter and activate activity of RNA polymerase
Regulate gene expression through transcription Often allosterically regulated by small molecules, such as cAMP |
|
|
Repressors
|
Proteins that bind DNA close to promoter and repress activity of RNA polymerase
Regulate gene expression through transcription Often allosterically regulated by small molecules, such as cAMP |
|
|
Operon
|
A sequence of bacterial DNA containing an operator, promoter and genes that contribute to single prokaryotic mRNA
The genes outside the operon may code for activators and repressors |
|
|
Lac Operon
|
Codes for enzymes that allow E. coli to import or metabolize lactose when glucose is not present in sufficient quantities
Lack of glucose in cells, leads to high levels of cAMP, which binds and activates import/metabolism of lactose Lac repressor protein is inactivated by presence of lactose in cells Lactose induces transcription of lac operon only when glucose is not present |
|
|
Primary transcript
|
Initial mRNA nucleotide sequence arrived at through transcription (AKA pre-mRNA or hnRNA)
|
|
|
Post-transcriptional processing
|
1. Addition of nucleotides
2. Deletion of nucleotides 3. Modification of nitrogenous bases |
|
|
5'-cap
|
5' end is capped
Attachment site in protein synthesis Protection against degradation by exonucleases |
|
|
Poly A tail
|
3' end is polyadenylated
Protects against degradation by exonucleases |
|
|
Introns
|
Non-coding sequence of DNA and RNA
|
|
|
Exons
|
Coding sequence of DNA and RNA
|
|
|
snRNPs
|
Enzyme-RNA complexes (small nuclear ribonucleoproteins) that recognize nucleotides sequences at end of introns
Associate with proteins to form spliceosome complex |
|
|
Splicing
|
Excision of introns from RNA by and splicing together of exons from RNA by snRNPs to form single mRNA strand that codes for polypeptide
Occurs in nucleus Introns remain in nucleus and are degrated Exons exit nucleus to be translated |
|
|
Denatured DNA
|
DNA molecule exposed to heat, high concentration of salt solutions or high pH solution
H-bonds connecting 2 strands of DNA are disrupted and separated DNA prefers to be double stranded and will look for a complementary partner to spontaneously associate into double-stranded DNA |
|
|
Nucleic Acid Hybridization
|
Process of forming various double stranded combinations:
1. DNA-DNA 2. DNA-RNA 3. RNA-RNA Enable identification of nucleotide sequences by binding a known sequence with an unknown sequence |
|
|
Restriction enzymes (AKA restriction endonucleases)
|
Digest nucleic acid only at certain nucleotide sequences along chain (restriction site)
Cleave strand unevenly, leaving complementary single stranded ends (sticky ends) Restriction site is typically palindromic, 4-6 nucs long |
|
|
Vector
|
Also known as plasmid
Used to place DNA within bacteria |
|
|
Screening DNA libraries
|
Identifying desired clones from various possibilities existing in DNA library:
1. No vector 2. Vector, no DNA fragment 3. Vector + DNA fragment page 41 |
|
|
Complementary DNA (cDNA)
|
DNA reverse transcribed from mRNA
Lacks introns Adding DNA polymerase to cDNA produces double strand of desired DNA fragment |
|
|
PCR
|
Polymerase chain reaction: Target DNA is denatured and mixed with complementary primers. Specialized polymerase replicates target DNA
Fast way of cloning DNA Uses specialized polymerase 3 steps: 1. Denaturing 2. Annealing 3. Extension/amplification |
|
|
Anneal
|
primers hybridize (bind) to complementary ends of DNA strands
|
|
|
Southern Blotting
|
Technique used to identify target fragments of known DNA sequence in large population of DNA
Identifies specific sequences of DNA by nucleic acid hybridization 1.DNA is cleaved into restriction fragments 2. Fragments are separated according to size via gel electrophoresis 3. DNA fragments are denatured via alkalination 4. Gel is transferred onto membrane 5. Radio-labeled probe with complementary nucleotide sequence to target fragment is added to membrane 6. Membrane is exposed to radiographic film |
|
|
Northern Blotting
|
Just like southern blot except it identifies RNA fragments
|
|
|
Western Blotting
|
Detects a particular protein in a mixture of proteins with antibodies
|
|
|
RFLP (Restriction fragment length polymorphisms)
|
Identifies individuals as opposed to identifying specific genes
Used to identify criminals |
|
|
Start codon
|
AUG = methionine
Signal beginning to protein synthesis - translation |
|
|
Stop codons
|
UAA, UAG & UGA
Signal end to protein synthesis - translation |
|
|
A polypeptide has 100 AA's. How many possible AA sequences are there for this polypeptide?
|
20^100.
# things (codons or AA's) ^ # positions = # of possible combinations |
|
|
Anticodon
|
tRNA contains set of nucleotides that is complementary to codon
|
|
|
Ribosome
|
Site of translation
Composed of small subunit and large subunit made from rRNA and many separate proteins Measured by sedimentation coefficients Prokary: 30s and 50s, total 70s Eukary: 40s and 60s, total 80s Manufactured in the nucleolus Small & large subunits exported separately to cytoplasm |
|
|
P site
|
Peptidyl site
Site at which tRNA possessing 5'-CAU-3' anticodon sequesters amino acid methionine at small subunit Signal for large subunit to join (makes the initiation complex) |
|
|
Initiation
|
Process of joining of large subunit to small subunit of ribosome
|
|
|
Elongation
|
Adding of amino acids to form polypeptide chain
|
|
|
A site
|
Aminoacyl site
tRNA with corresponding amino acid attaches to this site |
|
|
Translocation
|
Step in elongation
Ribosome shifts 3 nucleotides along mRNA toward 3' end. mRNA is always read 5' to 3'. |
|
|
E site
|
tRNA that carried methionine moves to this site, where it can exit ribosome
|
|
|
Termination
|
Translation ends when a stop or nonsense codon is reached
Polypeptide is freed from tRNA and ribosome when release factor binds to A site, allowing water molecule to add to end of polypeptide chain Ribosome breaks apart into subunits to be used again |
|
|
Post-translational modifications
|
1.Sugars, lipids or phosphate groups may be added to amino acids on the polypeptide
2. Polypeptide may be cleaved in one or more places 3. Separate polypeptides may join to form quaternary structures of proteins |
|
|
Places translation can take place:
|
1. Free floating ribosome in cytosol, producing proteins that function in cytosol
2. Ribosome may attach to rough ER during translation and inject proteins into ER lumen, which are destined to become membrane bound proteins of nuclear envelope/ER/golgi/lysosome/plasma membrane/secreted from cell |
|
|
Signal recognition particle (SRP)
|
Recognizes 20 AA sequence near front of polypeptide. It carries entire ribosome complex to a receptor protein on ER
|
|
|
Gene mutation
|
Alteration in DNA nucleotide sequence in single gene
|
|
|
Chromosomal mutation
|
Structure of chromosome is changed
|
|
|
Mutagens
|
Induced mutations due to physical or chemical agents
Increases frequency of mutation above frequency of spontaneous mutations |
|
|
Point mutation
|
Alteration of single base pair of nucleotide in double stranded DNA
|
|
|
Base-pair substitution mutation
|
Type of point mutation
Results when one base pair is replaced by another |
|
|
Missense mutation
|
Point mutation
Base pair mutation that occurs in amino acid coding sequence of a gene May or may not alter amino acid sequence of protein May or may not have serious affects on function of protein |
|
|
Insertion or deletion mutation
|
Point mutation
Insertion or deletion of a bp May result in a frameshift mutation |
|
|
Frameshift mutation
|
Results when deletion or insertion mutation is not in multiples of 3
Entire sequence will be shifted Often result in complete nonfunctional proteins |
|
|
Nonsense mutation
|
Bp substitution, deletion or insertion creates a stop codon
Prevent translation of functional protein, resulting in truncated nonfunctional protein |
|
|
Chromosomal deletions
|
Portion of chromosome breaks off or lost during homologous recombination or crossing over events
|
|
|
Chromosomal duplications
|
DNA fragment breaks free of one chromosome and incorporates into a homologous chromosome
|
|
|
Translocation
|
Segment of DNA from one chromosome is inserted into another chromosome
|
|
|
Inversion
|
Orientation of section of DNA is reversed on a chromosome
ABCDEFG --> CBADEFG |
|
|
Transposable elements
|
Also known as transposons
DNA segments that can excise themselves from a chromosomes and reinsert themselves at another location |
|
|
Transposition
|
Mechanisms by which somatic cell of multicellular organism can alter is genetic makeup without meiosis
|
|
|
Forward mutation
|
Changes organism even more from original state
|
|
|
Backward mutation
|
Reverts organism back to original state (wild type)
|
|
|
Cancer
|
Unrestrained and uncontrolled growth of cells
|
|
|
Oncogenes
|
Genes that cause cancer
Mutated from proto-oncogenes via mutagens (UV radiation, chemicals, random mutations |
|
|
Carcinogens
|
Mutagens that cause cancer
|
|
|
Histones
|
Globular proteins around which DNA is tightly wrapped (DNA not being used)
Basic |
|
|
Nucleosome
|
8 histones wrapped in DNA
|
|
|
Chromatin
|
Entire DNA/protein complex (including very small amount of RNA)
1/3 DNA 2/3 Protein Small amount RNA Net positive charge at the normal pH of cells because of basicity of histones |
|
|
Chromosome
|
Chromatin associated with each of 46 DNA molecules
Each contains hundreds or thousands of genes 46 inside nucleus of human somatic cell (diploid) |
|
|
Homologues
|
Partner chromosome that codes for same trait
One from mom and one from dad |
|
|
Diploid
|
Cell that contains homologous pairs of chromosomes
46 chromosomes |
|
|
Haploid
|
Cell that does not contain homologous pairs of chromosome
23 chromosomes |
|
|
Interphase
|
G1, S & G2 phases collectively
|
|
|
G1 phase
|
Cell has just split
Begins to grow in size Producing organelles and proteins G1 checkpoint at the end of G1 allowing cell to advance to S phase. If doesn't pass checkpoint G1, goes to G0 |
|
|
Chromatids
|
At the end of S phase, cell contains same number of chromosomes, except each chromosome is made up of identical sister chromatids
|
|
|
G2 phase
|
Cell prepares to divide
Growth phase G2 checkpoint checks for mitosis promoting factor (MPF) - if high enough, mitosis phase begins |
|
|
M phase
|
Mitosis = nuclear division without genetic chance
There is a M checkpoint which triggers G1 phase |
|
|
4 stages of mitosis:
|
1. Prophase
2. Metaphase 3. Anaphase 4. Telophase Results in genetically identical daughter cells |
|
|
Prophase
|
1. Condensation of chromatin into chromosomes
2. Centrioles (the two tubes) move to opposite end of cell 3. Nucleolus and nucleus disappear 4. Spindle apparatus begins to form, consisting of aster 5. Kinetochore microtubules grow from centromeres 6. Spindle microtubules connect 2 centrioles |
|
|
Aster
|
Microtubules radiating from centrioles
|
|
|
Centromeres
|
Group of proteins located at center of chromosome (at the chromosome's waist)
|
|
|
Kinetochore
|
Structure of protein and DNA located at centromere of joined chromatids of each chromosome
|
|
|
Metaphase
|
Chromosomes align along equator of cell
|
|
|
Anaphase
|
1. Sister chromatids split at attaching centromeres and move toward opposite ends of cell (disjunction)
2. Cytokinesis = actual seperation of cellular cytoplasm due to constriction of microfilaments about center of cell (end of anaphase) |
|
|
Telophase
|
1. Nuclear membrane reforms
2. Reformation of nucleolus 3. Chromosomes decondense 4. Cytokinesis continues |
|
|
Meiosis
|
Double nuclear division
Produces 4 haploid cells (gametes) |
|
|
Gametes
|
Germ cells
1. Spermatogonium 2. Oogonium |
|
|
Spermatogonium and Oogonium undergo meiosis:
|
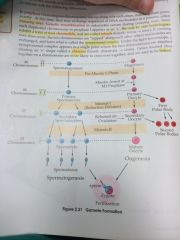
spermatogonium -> primary spermatocyte -> 2 secondary spermatocytes -> 4 spermatids -> 4 spermatozoa
Oogonium -> Primary Oocyte (+1 polar body) -> secondary Oocyte (+1 or 2 polar body) -> Mature Oocyte |
|
|
2 phases of meiosis:
|
1. meiosis I = similar to mitosis
2. meiosis II |
|
|
Meiosis I differs from mitosis:
|
Prophase I homologous chromosomes line up along side each other (total of 4 chromatids = tetrads)
may exchange sequences of DNA = crossing over (AKA genetic recombination) |
|
|
Crossing over
|
Process of exchanging DNA sequences between homologous chromosomes during Prophase I of Meosis I
|
|
|
Chiasma
|
X-shape of single point where 2 chromosomes are attached during crossing over events
|
|
|
Linked genes
|
Genes located close together on a chromosome
More likely to cross over together |
|
|
Metaphase I
|
Homologues remain attached
Move to metaphase plate and align as tetrads (unlike in mitosis, where single chromosome align) |
|
|
Anaphase I
|
Separates the homologues from their partners
|
|
|
Telophase I
|
Nuclear membrane may or may not form
Cytokinesis may or may not occur In humans, both do happen New cells, after cytokinesis, are halpoid (23 replicated chromosomes) |
|
|
Secondary spermatocyte or oocyte
|
Haploid (23 replicated chromosomes) daughter cells resulting from Meiosis I
|
|
|
Meiosis I
|
1. Prophase 1
2. Metaphase 1 3. Anaphase 1 4. Telophase 1 Reductive division |
|
|
Meiosis II
|
1. Prophase II
2. Metaphase II 3. Anaphase II 4. Telophase II Under light microscope, appears like normal mitosis Final products are haploid gametes with 23 chromosomes 4 sperm cells are formed Single ovum is formed (telophase II produces one gamete and a second polar body) |
|
|
Nondisjunction
|
During anaphase I (primary) or II (secondary) of meiosis
If centromere of any chromosome does not split |
|
|
Primary nondisjunction
|
One cell will have 2 extra chromatids, complete extra chromosome
Other cell will be missing a chromosome |
|
|
Secondary nondisjunction
|
One cell will have an extra chromatid
Other cell will lack a chromatid |

