![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
24 Cards in this Set
- Front
- Back
|
∆Gfolding |
∆Gfolding = Gfolded - Gunfolded |
|
|
Protien stabilization forces |
H-bonding Hydrophobic interactions Van der Waals interactions Disulfide bonds Metal binding |
|
|
H-bonding protein stabilization |
-Backbone amide and other functional groups are donors/acceptors -H-bonds w/ H2O (folded and unfolded) -H-bonds between protein atoms (folded) -H-bonds between backbone -NH and -CO groups help determine 2' structures |
|
|
Hydrophobic protein stabilization |
-Minimize exposure of non-polar groups to H2O -MAJOR driving force in protein folding |
|
|
Van der Waals protein stabilization |
-Maximized in folded tertiary (3') structure |
|
|
Disulfide protein stabilization |
-Intra & inter chain -2 oxidized Cys residues -Covalent bond |
|
|
Metal binding stabilization |
-Can stabilize under reducing conditions (intracellular) |
|
|
Conformation |
Spatial arrangement of atoms in a protein or any part of a protein |
|
|
Native protein |
Protein in its functional, folded conformation |
|
|
Peptide bond conformation |
-Rigid and planar -Amide N and and cabonyl O exhibit resonance -Discovered by Pauling and Cory |
|
|
Peptide rotation |
Peptide bond cannot rotate b/c partial double bond character N-Calpha (phi) C-Calpha (psi) +/- 180 degree rotation |
|
|
Rmachandran plot |
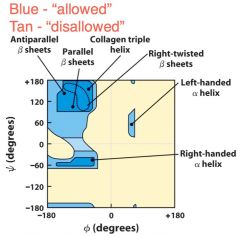
Plot of phi vs psi for amino acid residue showing allowed values of each |
|
|
Dihedral angles |
Angles that allow peptide rotation |
|
|
a-helix |
-Right handed, N-C -1 turn = 5.4 a, 3.6 amino acid residues -R groups on outside, h-bonds inside -Bulky/charged residues must be separated -amphiphilic, helicies segregate polar and nonpolar (for instance: left side polar, right side non-polar) |
|
|
Which AA has greatest tendency to form an a-helix? |
Alanine |
|
|
Helix dipole |
Forms b/c H-bonds transfer dipole of peptide bonds resulting in a + N terminus and a - C terminus |
|
|
B-sheets |
-Zig-zag B-strands align to form sheets -Sheets stabilized by H-bonding between Amino and Carbonyl of other strands -Sheets can run parallel (6.5 a per unit) or antiparallel (7 a per unit) |
|
|
B-turns |
-Form in places where direction changes -link successive a-helices of B-strands -180 degree turn involving 4 aa residues -1st residue carbonyl oxygen h-bonds w/ H of the last residues N -Gly and Pro often occur |
|
|
Fibrous proteins |
-Elongated with short repeating motifs -Lack defined tertiary structure -Provide strength --> fibrins/sheets -a-Keritin --> hair, nails, feathers |
|
|
Collagens |
-Fibrous proteins -Limited # aa types -~35% Gly, 11% Ala, 21% Pro (+ 4-Hyp) (4-hydroxyproline) -Lack defined tertiary structure -2' structure = collagen helix -4' structure = coil of 3 helicies |
|
|
Globular proteins |
-Sequences and structures are complicated -Structures determined via x-ray cryst., NMR, (& electron microscopy) -Multiple 2' structure elements -Helices and strands comprise core -Turns are on surface |
|
|
-Motif, fold, or supersecondary structure |
Recognizable folding pattern involving two or more elements of secondary structure and the connections between them |
|
|
4 classes of motifs |
-all a -all B -a/B (B-a-B loop repitition) -a + B: alpha and beta domains seperated |
|
|
Intrinsically Disordered Proteins |
-Lack hydrophobic core -High densities of charged amino acid residues - >25% of proteome -No distinct structure |

