![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
94 Cards in this Set
- Front
- Back
|
basic principles of pharmaceutical microbiology
|
Drug safety, laboratory diagnosis, development and evaluation of antimicrobial agents, disinfection, pharmaceuticals produced by microorganisms, detection of mutagenic/carcinogenic activity
|
|
|
benefits associated with microorganisms in pharmacy
|
Abx
steroids therapeutic enzymes polysaccharides recombinant DNA tech vaccines knowing Abx vitamin and AA conc detect mutations/carcinogens |
|
|
Problems associated with microorganisms in pharmacy
|
contamination of medicine
infectious agents risk of pyrogenic rxns reservoir of Abx resistant genes |
|
|
Virus
|
size: 20-400 nm
genome: ds/ss, linear/circular, DNA/RNA architecture: nucleic acid enclosed within a capsule/capsid made of proteins metabolism: within host cells |
|
|
Viroid
|
size: smallest known pathogens; 120-475 nucleotides
genome: ssRNA w/o associated protein architecture: just RNA metabolism: replicate in nucleus or chloroplasts of plant cells |
|
|
Prions
|
size: <100 nm
genome: lack of nucleic acid architecture: host-derived glycoproteins; major prion protein (PrPsc); can fold in multiple structurally distinct forms; mis-folded protein metabolism: slow replication (polymers may split into two smaller polymers, which results in two infectious polymers capable of further lengthening) |
|
|
Bacteria
|
size: Ranges from 0.5-5.0 micrometers, varies
genome: essential and nonessential DNA, though varies architecture: gram(+), gram(-), shape metabolism: varies, some replicate using free-floating DNA, from another bacteria via pili, or some self-replicate |
|
|
Bacterial architecture: Phenotypic classification
Biotyping |
epidemiologic purposes: biochemical markers: fermentation, presence of hydrolytic enzymes
|
|
|
Bacterial architecture: Phenotypic classification
Serotyping |
presence of unique antigens
|
|
|
Bacterial architecture: Phenotypic classification Antibiogram
|
patterns classify abx resistance
|
|
|
Bacterial architecture: Phenotypic classification Phage typing
|
classifying type of phage
|
|
|
Fungi
|
size: variable
genome: DNA architecture: eukaryotes distinct from plants and animals; multinucleate or multicellular, thick cell wall with chitin, grow like thread-like filaments “hyphae” or as ovoid or spherical, single cells (yeast) metabolism: morphogenesis (ability to transform from saprobic filamentous mould to unicellular yeast in host). they can reproduce sexually/asexually |
|
|
Protozoa
|
size: single cell
genome: DNA architecture: sporozoa (intracell), flagellates (move with flagella), amoebae (move with pseudopodia), ciliates (move with cilia) metabolism: intracellular or extracellular, free living |
|
|
Virus Morphology
|
Morphology: comparing size/shape/traits
Examples: capsomeres, glycoproteins, bacteriaphage’s head, sheath, and tail fiber |
|
|
Bacterial Morphology
|
Examples: Shape (cocci, spiral, bacilii), gram (+) or (-)
|
|
|
Fungi Morphology
|
Example: changing from a yeast form to a capsule form
|
|
|
Vaccine preventable Diseases
|
-measles, rabies, hep A & B, varicella zoster virus, smallpox, human papillomavirus, rubella, mumps, influenza A & B, poliovirus, rotavirus (Mr. hab VS. hrmiab pv)
-parainfluenza virus for DOGS |
|
|
Gram (+) Cell wall
|
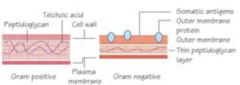
somatic antigens, outer membrane protein, thin peptidoglycan layer
|
|
|
Gram (-) Cell wall
|
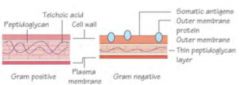
large peptidoglycan layer, teichoic acid
|
|
|
Diagnosis: Microscopy
|
old fashion, staining plates
|
|
|
Diagnosis: Culture
|
less ability to trace abx resistance, broth medium or plate, identification by biochemical or serological tests on pure growth from single colony, help ID pathogen
|
|
|
Diagnosis: Sensitivities
|
by disc diffusion methods, breakpoints or MICs, serodiagnosis, DNA technologies
|
|
|
disinfection
|
process of removing microorganisms, including pathogens from the surfaces
|
|
|
preservation
|
use of preservatives to prevent microbial spoilage of the product, must not be toxic
|
|
|
antisepsis
|
destruction or inhibition of microorganisms on living tissues to limit infection
|
|
|
antiseptics
|
must not be toxic or irritating for these tissues; to reduce microbial population
|
|
|
sterilization
|
any process that eliminates (removes) or kills all forms of life
|
|
|
Physical Sterilization
|
sunlight, heat, vibration, radiation, filtration
|
|
|
chemical sterilization
|
liquid, gaseous
|
|
|
physiochemical sterilization
|
combo of the two
|
|
|
MBC
|
minimal bactericidal concentration: lowest concentration of antimicrobial that will prevent growth of microorganism after subculture onto antibiotic-free media
|
|
|
MIC
|
minimal inhibitory concentration: lowest concentration of a compound which will inhibit the visible growth of microorganism after overnight cultivation
|
|
|
Drug discovery using phage display
|
pool peptides - take target you want phage to bind (ex cell, receptor, etc), incubate it in presence of target, wash to remove the unbound leaving only phages w/affinity to target, ellute and amplify using E coli. Repeat a few times to remove phages w/lower binding affinities.
|
|
|
Ames Test
|
-screen chemicals for potential carcinogenicity or anti-cancer cmpd
-large # cmpds can be screened by examining ability to induce mutagenesis in specially constructed mutant derived from Salmonella enterica -process: bacteria grow in presence of liver extract and His requiring salmonella→ mutagen added → plated w/o His and incubated → plates examined for control (no growth) and growth (added chemical cause inc in His revertants - mutagenic) TL;DR: See if a cmpd causes mutations in salmonella. His helps growth--use little His to see if adaptation/mutation occurs |
|
|
bacteriostatic
|
inhibits growth of bacteria
|
|
|
bactericidal
|
kills bacteria
|
|
|
Pharmaceuticals produced by microorganisms
|
-antibiotics
-dextrans (polysaccharides produced by lactic -acid bacteria, L dextranicus or L mesenteroides, following growth on sucrose) -vitamins (B1, B12) -amino acids -organic acids (citric, lactic, gluconic) -iron chelating agents (siderophores) -enzymes (neuraminidase, streptokinase) |
|
|
eubiosis
|
good co-existence of host and microflora - symbiosis. protection of intestinal mucosa, antagonistic effect on undesired microorganism, maturation and stim of immune system, nutrient digestion, vitamin synthesis, protein synthesis
|
|
|
dysbiosis
|
bad: damage to intestinal epithelium, gut wall thickening, reduced resorption of nutrients, toxic metabolic substances, decomp and inc gas prod (CH4, H2S, CO2), weaken immune system, immune rxn, accelerate cell turnover
|
|
|
HMP
|
Human microbiome project by NIH to assess how changes in microbiome correlate w/human disease
aims: create reference catalog of microbial genome sequences, large cohort studies to survey microbiome across healthy human body, correlation of changes in microbiome w/disease, computational tools to analyzing microbiome metagenomic sequence data |
|
|
characterizing microbiota using culture based approach
|
inability to detect uncultivable microorganisms
|
|
|
characterizing microbiota using culture independent methods
|
-16S rRNA as phylogenetic marker (specific to each bacteria)
-whole genome sequencing: high throughput sequencing methods (Illumina) offer read lengths that enable classification of sample microorganisms - METHOD OF CHOICE. better for phylogenetic resolution and allow for ID of unknown bacteria not detected w/microarray approaches |
|
|
Factors influencing origin/development of microbiota
|
depend on inoculum - intrauterina contamination due to translocation from mother’s intestinal microbiota
mode of delivery - vaginal v. C section type of feed - breast v formula weaning food practices use of antimicrobials familial environment |
|
|
stomach CFU
|
0-10^2
lactobacillus, candida, streptococcus, h pylori, peptostreptococcus |
|
|
duodenum CFU
|
10^2
streptococcus, lactobacillus |
|
|
proximal ileum CFU
|
10^3
streptococcus, lactobacillus |
|
|
distal ileum CFU
|
10^7-10^8
clostridium, streptococci, bacteroides, actinomycineae, corynebacteria |
|
|
jejunum CFU
|
10^2
streptococcus, lactobacillus |
|
|
colon CFU
|
10^11-10^12
bacteroides, clostridium groups IV and XIV, bifidobacterium, enterobacteriaceae |
|
|
irritable bowel syndrome (IBS)
|
-common GI disorder due to alterations in gut microbiotachronic recurring episodes of abdominal pain asso. w/altered bowel habit in absence of organic disease, sensation of bloating w/ and w/o visible abdominal distension, increasing trait anxiety, and several extraintestinal sx
-sx: ab pain, diarrhea, constipation, behavioral changes |
|
|
inflammatory bowel disease (IBD)
|
-due to defects in innate immunity (dec AMP, defect in mucosal barrier, abnormal mucin expression), autophagy (mutation in loci - granuloma form), phagocytosis (altered phagosome function)combined w/proinflammatory response and mucosal damage lead to dysbiosis and inflammation, ulceration/fibrosis, metaplasia/cancer
-sx: diarrhea, weight loss, ab pain, cramping, GI bleeding, obstructions |
|
|
prebiotic
|
-non living, non digestible form of fiber or carbs
-only metabolized by gut bacteria and NOT human host - role is to nourish intestinal bacteria -ideal prebiotic: not hydrolyzed or abs in upper GI, selective for beneficial bacteria in colon, alter colonic microbiota towards healthier composition, stimulate growth of intestinal bacteria w/health and well being, help inc abs of minerals (Ca, Mg), favorable immune effect, provided resistance against infection -good substrates for commensal bacteria -affect: immune function, colon cancer, microflora modification, BG, lipid metab, mineral abs, laxation, diarrhea, IBD, IBS |
|
|
probiotic
|
-live bacteria, active bacterial cultures w/beneficial host effects to fight bacteria in gut
-features: non-pathogenic, non-toxic, non allergic, can survive in metabolizing upper GI, resistant to low pH, organic acid, bile juice, saliva, gastric acid, stable and capable of remaining viable for long periods, can modulate immune response, provide resistance to disease via improved immunity or production of antimicrobial substance in gut, good adhesion to GI tract and influence on gut mucosal permeability, antagonistic against carcinogenic organism, health benefit again GI disorder, able to be sold (stable) -effects: aid in lactose digestion, resistance to enteric pathogen, anti colon cancer, anti HTN, small bowel bacterial overgrowth, immune system modulation, blood lipid, heart dz, urogenitc infection, hepatic encephalpathy |
|
|
synbiotic
|
combo of the two
|
|
|
Clostridium deficile
|
-hazard level urgent
-anaerobic spore forming bacillus cause pseudomembranous colitis, toxic megacolon sepsis, death. -Disrupts tight jxns to cause diarrhea -fecal oral transmission via contaminated environ, hands of healthcare personnel -pass thru GI, colonize intestine and secrete toxin A (tcdA) and B (TcdB) |
|
|
tcdA
|
bind and cytoskeletal change that alter tight junction to loosen epithelial barrier or produce inflamm mediators
|
|
|
tcdB
|
cytotoxic and induce release of various mediators, phagocytes, mast cells
|
|
|
carbapenem resistant enterobacteriaceae (CRE)
|
hazard level urgent
|
|
|
drug resistant Neisseria gonorrhoeae (cephalosporin resistance)
|
hazard level urgent
|
|
|
bacteria of class Enterobacteriaceae
|
gram (-)
escherichia/shigella, salmonella, yersinia, proteus, klebsiella - 41 genera |
|
|
Major virulence factors of Enterobacteriaceae
|
enterotoxins: heat labile or stable, verotoxin (VT1, VT2, VT2e), cytotoxic necrotizing factor (CNF1, CNF2), endotoxin, exotoxins
|
|
|
ETEC
|
-enterotoxigenic E. coli
-cause of bacterial diarrheal illness, traveler's diarrhea -virulence factor: heat labile (LT1 and LT2) and heat stable (ST) -coded for by mobilizable plasmid, effect similar to cholera → result in electrolyte and water efflux into intestinal lumen w/little inflammation |
|
|
EHEC
|
-enterohemorrhagic E coli
-affect large bowel -related to STEC -virulence factor: shiga toxin (A/B - A subunit bind 28s rRNA and shut down protein synthesis that cause to cell death) enter blood stream and bind Gb3 receptor on renal tissue and LEE pathogenicity island (pedestal formation) |
|
|
EPEC
|
-enteropathogenic E coli
-cause of infant diarrhea in impoverished countries - n/v, fever, non-bloody stool -effacement of microvilli result in loss of absorptive property of intestine, inflammation, and watery diarrhea -nature of virulence factor uncertain, possibly: cortactin, cytokeratin 8, cytokeratin 18, dynamin, GRB2, LPP, SHC, vinculin, zyxin |
|
|
UPEC
|
-uropathogenic E coli
-adhesins required for colonization - damage bladder (inflammation) but does not typically involve kidney -local rxn attributed to endo and exotoxins -virulence factor: P1 pili (attach to bladder cell or survive/grow in urine) |
|
|
NMEC
|
-neonatal meningitis
-affect bloodstream |
|
|
C. diff
|
route: fecal oral via contaminated environment or healthcare professional hands
sx: pseudomembranous colitis, toxic megacolon sepsis, death MOA: disrupt tight junctions to cause diarrhea |
|
|
E. coli
|
route: carrier (cow) → contaminated meat, food, beverage → primary infection (of consumer) → secondary infection (ie if breast feeding)
-spread through fecal matter EPEC - sx: fever, n/v, non bloody stool EHEC - sx: stomach muscle spasm, diarrhea (can be bloody), fever, vomit UPEC - sx: UTI |
|
|
shigella
|
-Subset of E.coli, produces enterotoxins, shiga toxins, facultative intracellular, large virulence plasmid-absolutely associated with disease
-sx: diarrhea, fever, stomach cramp 2 days after exposure |
|
|
salmonella
|
-From chicken eggs/food, bacteria travels to small intestine and adheres to the lining to reproduce.
-sx: diarrhea, fever, ab cramp 12-72 hr after infection |
|
|
klebsiella
|
-Enters the respiratory tract, can be spread through vents, catheters, wounds, people
-sx: pneumonia |
|
|
Campylobacter jejuni
|
Found in feces and raw chicken. xmit to weak immune sys pt's. curved, helical shaped, gram(-), grows best at 37 to 42C
-sx: diarrhea, cramp, ab pain, fever |
|
|
Pseudomonas aeruginosa
|
route: ppl in hospital or w/weak immune system - found in soil and water
-gram(-), rod, motile, fruity odor, greenish-blue color, many virulence factors. sx: CNS, local (skin through burns/wounds), heart, respiratory, and urinary tract infections |
|
|
Acinetobacter baumannii
|
route: soil and water → infections in ICU or housing of ill pts.
-colonize pt w/o infection or symptoms, especially on skin, in tracheostomy sites or open wounds, pt with weak immune system, chronic lung, or DM. sx: Causes respiratory tract infections (pneumonia) and UTI’s. |
|
|
stentorophomous maltophilia
|
route: aqueous habitats (animal, food, water source)
-resistant to most abx |
|
|
Neisseria gonorrhoeae
|
coccus
human-specific pathogen |
|
|
LPS
|
-lipopolysaccharide
-impedes destruction of bacteria by serum components and phagocytic cells; may be involved in adherence or antigenic shifts that determine the course of infection |
|
|
capsule
|
serum resistance, capsule protects against complement mediated killing
|
|
|
enterotoxins
|
affect the permeability of small intestine.
Ex. shiga toxin, other toxins |
|
|
phase variation of antigens
|
pilli, flagella, capsule, O antigen, protects from antibody-mediated cell death
|
|
|
sequestration of growth factors
|
iron: production of siderophores (iron-chelators) called enterobactin, aerobactin
|
|
|
T2SS
|
-multicomponent machines that use two-step mechanism for translocation
-step 1, precursor effector protein is translocated through the inner membrane by the Sec translocon or the Tat pathway -in the periplasm, the protein is translocated by the T2SS through the outer membrane -the T2SS consists of 12-16 protein components that are found in both the bacterial membranes, the cytoplasm and periplasm |
|
|
T3SS
|
-injectisomes
-single-step secretion used by many plant and animal pathogens, including T3SSs deliver effector proteins into the eukaryotic host cell cytoplasm -genetically, structurally and functionally related to bacterial flagella -composed of more than 20 different proteins, which form a large supramolecular structure crossing the bacterial cell envelope |
|
|
T4SS
|
versatile systems found in Gram(-) and Gram(+) that secrete a wide range of substrates from single proteins to protein-protein and protein-DNA complexes
|
|
|
LPS structure
|
-carbohydrate: exposed at bacterial cell surface, very antigenic. varies between and within species
-lipid A portion: integral in bacterial outer membrane, constant among species, toxic portion of LPS (toxicity is mediated by interaction with innate immune system, TLRs |
|
|
TLR recognition
|
parts of the bacterial cell walls: LPS, unmethylated CpG DNA sequences, flagellin, peptidoglycan, lipoteichoic acid
|
|
|
flagella
|
“hair to move” that allow bacterial motility
|
|
|
monotrichous
|
flagella located just on one side
|
|
|
lophotrichous
|
many flagella located on both sides
|
|
|
amphitrichous
|
singe flagella located on both sides
|
|
|
peritrichs
|
many flagella located all over
|
|
|
pili
|
large fimbriae
|
|
|
fimbriae
|
-long structures that extend off surface of bacterium that mediate attached
-composed of pillin to bind carb residues of glycolipid or glycoprotein at host cell -initial first point of contact that allow transfer of DNA from donor to new cell |
|
|
afimbrial adhesins
|
-surface protein not structurally related to fimbriae, not structurally organized like pili with much variation in structure and function
-different proteins that enable bacteria to attach to host cell via glycoprotein, glycan, etc |

