![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
62 Cards in this Set
- Front
- Back
|
Levels of protein Structures |
4 Different Levels Primary, Secondary, Beta Pleated Sheet AKA Tertiary, and Quaternary |
|
|
The Primary Level of A protein Structure |
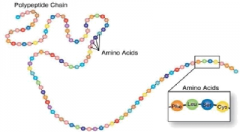
is a Sequence of amino acids in a polypeptide chain ( Amino acids are held together by Peptide Bonds ) |
|
|
The Secondary Level of a protein structure |
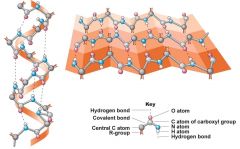
Folding based on amino acid interactions Alpha helix, and Beta Pleated sheet |
|
|
The Tertiay Level of a protein |
Further folding to form a 3D structure |
|
|
The quaternary structure level of a protein |
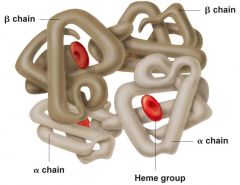
Interactions of two or more polypeptide cains to produce one protein ( not all proteins have a Quaternary structure) <---A human hemoglobin (Two Beta, Two Alpha chains |
|
|
Beadle and tatum ( 1940s) |
Worked with a muntant fungus, Neurospora They obtained strains that had genetic blocks of reactions essential to growth Complete medium- contained all essential nutrient components Minimal medium - lacking a nutritional component essential for growth Original Hypotheisis - One gene : one enzyme Changed to- ONe gene : one polypeptide |
|
|
Polypeptides |
ONE gene produces one polypeptide A polypeptide is a chain of amino acids joined by a covalent peptide cond |
|
|
Protein |
A protein is made from one or more polypeprides Proteins and enzymes and structually similar but have different functions Nearly all Enzymes are proteins , but not all proteins are enzymes |
|
|
Enzymes |
Nearly all Enzymes are proteins , but not all proteins are enzymes
|
|
|
Hemoglobin |
Is the oxygen- carrying protein in red blood cells they conatin 2 Alpha and 2 beta chains ( two ifferent polypeptides combining to equal one/ make one protein ) |
|
|
Sickle- cell anemia |
Controlled by a single gene with two alleles Encodes for a protein other than an enzyme A single nucleotide difference encodes a single amino acid difference, which causes the difference in hemoglobin and the shape of the red bood cells between normal and sickle cell individuals |
|
|
Types of Mutations |
Chromosome mutaions Gene mutaions |
|
|
Chromosome Mutation |
Change in chromosome number or structure |
|
|
Types of gene mutaion |
Gene mutations - lead to formation of a different alleles and is heritable change of a gene Forward vs. reverse spontaneous vs. induced - cause Gamic vs. somatic, where |
|
|
Forward vs. Reverse |
Forward mutation: changes away from the wild type allele Reverse mutation: Changes back to the wild type allele |
|
|
Gametic Vs. Somatic |
Gametic mutation : A mutation that happens in Gametes, It can be transferred to its offspring Somatic Mutation: A Mutation that happens in Somatic cells ( Body Cells ) Could be transmittd to offspring Only if they produce gametes |
|
|
Spontaneous Vs. Induced |
Spontanerous mutation - mutaion that happens in nature, Replication errors and Base modifications ATCG Induced Mutation - A mutaion induced by artificial factors, such as irradiation and chemical reactions |
|
|
Classification based on a Phenotypic effect |
A change in the DNA base sequence can cause a change in the final protein product A change in the final product will change phenotypes *Point mutations ( 3 types ) *Frame shift Mutaion |
|
|
Point Mutation |
Changes one base in the DNA sequence (3 types) Same sense - Specifies the same amino acid Mis sense- specifies a different animo acid Non sense - specifies a stop codon |
|
|
Frameshirft mutations |
Changes the reading frame an insertion or deletion of one or more bases |
|
|
Same Sence Mutation |
Mutation that changes a codon encoding one amino acid to another codon encoding the same amino acid ( changes the base sequence but not the amino acid sequence ) |
|
|
Mis sence mutation |
Sickle cell Mutations that changes a codon endocing one amino acid to another codon encoding a different amino acid ( changes the base sequence and the Amino acid sequence ) Mutation of one base causes incoporation of one different amino acis , wich results in a deformed protein |
|
|
non- sense mutation |
Mutation that changes a codon encoding one amino acid to another codon encoding a stop codon and signaling the end of a translation trucates translation and results in a sorter protein |
|
|
Detection of a mutation |
A gene mutation generally occus on only one allele at a gene locus In a haploid organism - there is immediater expression of the mutant allel In a Diploid Organism a recessive allele becomes a dominant allele ( can be detected in same generation ) recessive mutation a dominant allele becomes a recessive allele |
|
|
Origin of spontaneous mutations |
1.) DNA replication errors - Mispairing / mismatchinf of nuclotides
- Replication slippage
Most common in Fragile x , Huntingtons |
|
|
-Replication slippage
|
Causes 1.) Looping out of the DNA strand 2.) slippage of the DNA polymerase Results 1.) insertions or deletions 2.) frameshift mutations of amino acid insertions or deletions Most common in Fragile x , Huntington's |
|
|
Origin of spontaneous mutations 3 types
|
DNA relpication errors Base modification Oxidative damage |
|
|
Origin of spontaneous mutations Base mutations |
Chemical changes resulting in hydrogen bonding with non complemenary bases C-A T- G |
|
|
Origin of spontaneous mutations Oxidative damage |
by products of some normal cellular processes superoxides (02-) hydrocals (oh) exposure to high energy radiation ionizing radiation UV light |
|
|
Ultraviloet light |
can cause formation of tymine dimers , wich can destort the DNA |
|
|
UV dammage |
Can be reversed by nucleotide excision repair (NER) BUlky lesions are reconized and clipped out by products of the UUR genes DNA polymerase 1 and DNA ligase repair the gap |
|
|
Xeroderma Pigmentosum |
Recessive skin abnormaalities caused by sunlight ( UV ) damage NER system ( repair ) is not working May cause and produce skin cancer |
|
|
Origin of spontaneouse mutation |
Transposons Dna can move within or between genomes
|
|
|
Transposons |
They are widespread in plants Transposons Dna can move within or between genomes Also callled transposable genetics elements present in the genomes of all organisms can move to new locations and insert into the codinf regin or regulatory region of a gene, then result in mution of a gene can act as natually occuring mutagens |
|
|
Ac and DS elements |
Activator and dissociation discovered by Barbra McClintion she reported that bits of a maize genome could jump from place to place |
|
|
Transposons in bacteria |
Insertion secuence and transpospns |
|
|
Transposons in animals |
|
|
|
Transposons in Humans |
Lines : long interspersed elements Sines: Short Interspersed elements
|
|
|
Genetic regulation |
|
|
|
Operons |
A group of structual genes with a common prommoter ( regulator ) the genes within an operon are transcribed together they have a related biochemical function |
|
|
Genes within an Operon are transcribed together |
Polycistronic Mrna encodes multipule genes |
|
|
Prokaryotic Gene regulation |
|
|
|
inducible systems
|
they are normally turned off to express the gene they must be induced and turned on with the lac operon |
|
|
Repressible systems |
Are normally turned on they are expressed unless turned of using the trip operon |
|
|
The Lac Operon |
Produces three genes important for the metabolism of lactose( helps to induce this operon) when lactose is present the lac operon is turned on and proteins are produced to metabolize lactose with out lactose energy is conserved |
|
|
The Trip Operon |
Five adjacent genes involved in synthesis of tryptophan when tryptophan is present enzymes are not produced Tryptophan Helps to repress this operpn |
|
|
Three major differences in gene regulation Of eukaryotes vs. prokaryotes ( true for Eukaryotes )
|
|
|
|
HistoneModification
|
Histone proteins (the core of thenucleosome) have a positive charge and bind to DNAModification of histones _weakensthe interaction with DNA and allows transcription to occur
|
|
|
ChromatinRemodeling
|
Altering chromatin structure without chemical changes The SWI/SNF complex usesatp energy to _repositionnucleosomes,making DNA available for transcription
|
|
|
DNAMethylation
|
Methylation is tissuespecific Methylation is heritable,once established
Methylation generally hindersgene expression May represstranscription Addition of a methyl group, most often toCytosine |
|
|
Proliferation
|
abnormal cell growth and division
|
|
|
Metastasis
|
spread of cancer cells to other parts ofthe body
|
|
|
CancerAgenetic disease at the somatic level
|
Genomic alterations
Single-nucleotideSubstitution Chromosomerearrangements Chromosomaldeletions |
|
|
PotentialEffects of Chromosome Rearrangement
|
Deletions Eliminateor inactivate genes that control the cell cycle Inversions and Translocations Disruptgenes that suppress tumors Fusetwo genes to make cancer-causing proteins Aligngenes with different regulatory sequence |
|
|
ThePhiladelphia Chromosome
( chromosome rearrangement) |
|
|
|
Cancerand Epigenetics
|
|
|
|
Canceraffects many basic cellularfunctions
|
Cellcycle DNARepair systems Cell differentiation Cell migration Cell-to-cell contact Apoptosis (programmedcell death) |
|
|
NormalCell-Cycle Control and Checkpoints
|
Checklist
Cell sizeDNA integrityDNA replication Physiological conditions Spindle formation Spindle attachment |
|
|
Cyclins
|
regulate passage through the differentstages of the cell cycle
|
|
|
Cyclins dependent kinases
|
(CDKs) are enzymes that add phosphategroups toother proteins
CDKsfunction only in the presence of cyclinsActive cyclin/CDK complexes phosphorylate and activateother proteins that advance the cell through the cell cycle |
|
|
Cyclins |
Cyclinproteinsare made and destroyed in a precise pattern during the cell cycle
|
|
|
Twocategories of genes that causecancer when mutated
|
1.Proto-oncogenes
]Normalgenes that stimulates call division Initiates,facilitates, or maintains cell growth and division (e.g. cyclins) When mutated, it can become an oncogeneandcontribute to the _developmentof cancer _2.Tumor-suppressor genes Genesthat suppress unrestricted cell division |

