![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
72 Cards in this Set
- Front
- Back
|
What approach is used to amplify the sequence of interest? |
- Thereare more than 3000 million nucleotides in our genome - If wewant to analyze a single base, or even an exon (~150 bp),this is a tiny fraction of the DNA we would isolate from cells - Use amethod to amplify thesequence of interest: DNAcloning or PCR |
|
|
What approach is used to specifically recognize or detect the sequence of interest? |
- There are more than 3000 million nucleotides in our genome - If we want to analyze a single base, or even an exon (~150 bp), this is a tiny fraction of the DNA we would isolate from cells - Use amethod to specifically recognize (detect) the sequence of interest: Hybridization |
|
|
Explain: DNA Cloning |
- Amplificationof DNA using cells
- Methodof obtaining large amounts of a DNA sequence so that it can be studied or putto some use - Manyidentical copies - Usesbacterial cells, or less frequently, yeast cells |
|
|
How does DNA Cloning work? |
1. DNAsequence of interest is ligated (covalently joined) to a vectorDNA sequence that will help it replicate in the host cells - Maybe linear or circular - Includesa selection or screening system to help identify cells containing therecombinant DNA 2. Eachcell will take up only a single DNA molecule - Ifthere is a mixture of fragments this provides a method of sorting them intodifferent cells 3. InsertedDNA is amplified to a very high copy number, for two reasons: - Asingle bacterium will produce a huge number of identical bacterial cell clones,each with the same foreign DNA sequence - Somevector molecules will replicate within a bacterial cell to reach very high copynumbers per cell 4. Bacterialcells with different fragments are separated by spreading the transformed cellson agar plates - Willform well-separated cell colonies, each consisting of identical cellsoriginating from a single transformed cell 5. Asingle cell colony can be selected to start the growth of a large culture ofidentical cells with the sequence of interest - DNAwill be purified from this culture |
|
|
How does DNA Cloning look like? |
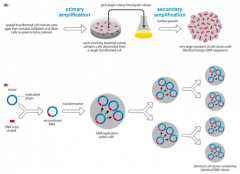
|
|
|
Define: Recombinant DNA |
- DNA sequence of interest is ligated (covalently joined) to a vector DNA sequence that will help it replicate in the host cells - Natural function is to protect bacteria from bacteriophages by cleaving foreign DNA - Bacteria’s own DNA is protected by methylation - e.g. EcoRI recognizes GAATTC - In E. coli, a methyltransferase recognizes this same sequence and methylates adenosine, preventing cleavage by the RE |
|
|
Define: Transformation |
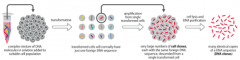
Transfersthe DNA molecules into the cell |
|
|
What are the three types of Vector Molecules? |
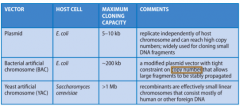
|
|
|
Define: Plasmids |
- Smallcircular dsDNA molecules that can replicate to very high copy numbers inbacterial cells - Geneticallyengineered to contain an antibiotic resistance gene - Aftertransformation, cells are grown on agar containing the antibiotic - Untransformedcells will die - Mayalso include a screening system to differentiate between cells with plasmidalone vs. cells with plasmid – target DNA recombinants |
|
|
What are components of Plasmid Vectors? |
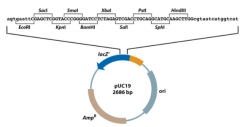
-LacZ - AmpR - Multiplecloning site - Ori |
|
|
Define: LacZ |
EncodesLacZ which will produce bluecolonies. Insertion of foreign DNA willinactivate the LacZ gene, producing white colonies. |
|
|
Define: AmpR |
Encodesresistance to the antibiotic ampicillin. |
|
|
Define: Multiple Cloning Site |
Containsmany restriction enzyme sites for DNA insertion |
|
|
Define: Ori |
Originof replication |
|
|
How do you make a Recombinant DNA? |
1. Eachfragment of interest needs to be covalently joined to a vector molecule, whichwill be transported into the host cell 2. Restrictionendonucleases are used to cut the DNA at defined places - Recognizespecific short sequence elements and cut the DNA on both strands within or nearthis site 3. Cleavageof target DNA and vector with the same RE produces complementary “sticky ends”that can hydrogen bond with each other DNAligase must be used to seal the fragments together |
|
|
Define: Restriction Endonucleases |

- Mostcommon recognize short palindromes - Cleavageoccurs at asymmetric positions within this sequence produces fragments withoverhanging 5’ or 3’ ends |
|
|
Define: DNA Library |
- Cloningcan be used to make a collection of DNA clones that contain all the DNAfragments from complex starting material - Ex. genomic DNA library |
|
|
Define: cDNA Library |
- Gene-centredDNA libraries can be made by isolating mRNA, and using reverse transcriptase tosynthesize cDNA that is cloned into plasmid vectors - cDNAlibraries will vary based on the cell type used! |
|
|
How are Genomic Libraries made? |
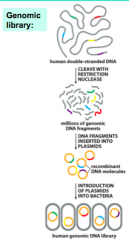
|
|
|
How are cDNA Libraries made? |
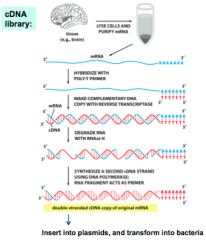
|
|
|
Comparison of Genomic Libraries and cDNA Libraries |
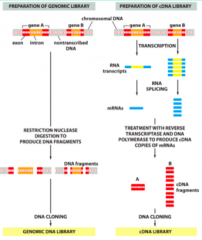
|
|
|
Explain: Expression Vectors |
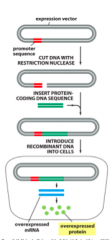
- Onegoal of DNA cloning may be to isolate a large quantity of protein fromthe DNA sequence - Canbe accomplished using expression vectors with highly active promoters - Cellsmay be bacteria, yeast, insect or mammalian cells |
|
|
What are the problems with using bacteria to express mammalian proteins? |
- Nopost-translational processing - Largemammalian proteins may misfold andform insoluble aggregates - Proteinmay be toxic to the bacteria - Mayhelp to use an inducible promoter |
|
|
Define: Fusion Vectors |
Insome cases, it may be desirable to fuse the protein of interest to anotherprotein or a small protein tag |
|
|
What are the advantages of using Fusion Vectors? |
- Fusionto some peptides or proteins increases yield and solubility in bacterial hostcells - Fusionto an affinity tag can be used to assist in purification of the recombinantprotein by affinity chromatography - e.g.Glutathione S-transferase (GST) or His6 tag - Fusionto a fluorescent tag allows the protein to be visualized or tracked in the cell |
|
|
How to prepare Fusion proteins? |
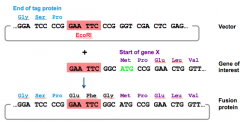
Whenpreparing fusion proteins, the reading frame of the two proteins must match inthe expression vector |
|
|
Define: PolymeraseChain Reaction (PCR) |
- Amethod of cloning DNA in vitro - Developedin mid 1980s |
|
|
What are the advantages of Polymerase Chain Reaction (PCR)? |
- Fast - Extremelysensitive - Allowsamplification from minute amounts of target DNA - Robust - Canamplify DNA from old, degraded, or embedded samples - Simplicity - Allowsfor parallel amplifications of DNA sequences from multiple starting samples |
|
|
What are the PCR Components? |
- Heat stable polymerase - Pairof single-stranded oligonucleotide primers - Reaction buffer and dNTPs |
|
|
Define: Heat Stable Polymerase |
Enzymethat is used to synthesize a small DNA sequence of interest within a complexstarting DNA (e.g. total genomic DNA) |
|
|
Define: Primers |

-Designedto bind to opposing DNA strands - Bindat sites flanking the DNA sequence of interest via sequence complementarity - Designedso the direction of synthesis of each new DNA strand is toward theother primer |
|
|
How does PCR Amplification work? |
1. Denaturationat 95°C, causing hydrogen bonds to break andDNA to become single stranded 2. Annealingat ~60 °C to allow primers to bind tocomplementary sequences 3. Extensionat ~72 °C to allow DNA polymerase to synthesizecomplementary strand beginning the free 3’-OH of one primer 4. Cycleis repeated 25 – 40 times - Witheach cycle, the amount of product ~doubles - Newlysynthesized DNA becomes a more likely target for primer binding &lification than original genomic DNA Endresult is millions of copies of the DNA sequence of interest |
|
|
What are the phases of PCR? |
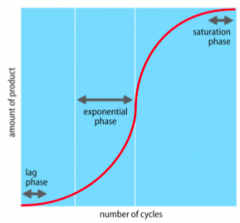
- In log phase, amountof PCR product increases gradually at first due to the complex pool of startingmaterial - Inthe exponential phase the reaction is operating at its highest efficiency - Productshave begunto accumulate and thereaction ~doubles these every cycle - Efficiencydecreases as the reaction components are consumed - Amountof product reaches its maximumin the saturation phase |
|
|
Define: Reverse Transcriptase |
Reversetranscriptase converts RNA to cDNA, which is used in a PCR reaction |
|
|
Explain: PCR Products |
- PCRproducts are analyzed on a gel - Nothighly quantitative - Exponentialphase usually completed - Goodfor detection of expression - Yes/No - Canbe used to identify transcript variants - Maybecome semi-quantitative if the number of cycles is reduced |
|
|
Explain: Quantitative Real-Time PCR |

- Performedon specialized PCR machines that detect the amount of product in each reactionin real-time as the reaction proceeds - Detectsthe amount of fluorescence signal in the sample, which is reflective of theamount of product (Ex. Sybr greencanbe included in the reaction and emits fluorescence when bound to dsDNA) - Oftenused with cDNA to quantify differences in gene expression in different samples - Theearlier a reaction enters the exponential phase, the more target sequencetherewas in the input =higher expression |
|
|
What are the disadvantages of PCR when compared to DNA Cloning? |
Shorterproduct lengths: - PCRis excellent for sequences up to 1 kb - Assize increases above that, it becomes more difficult - Cloningeasily allows for amplification of sequences several kb long - BACs& YACs: 100s of kb – Mb Lowerproduct amounts: - PCRsaturates after a limited number of cycles - Cloningallows for production of HUGE amounts of DNA |
|
|
Explain: Nucleic Acid Hybridization |
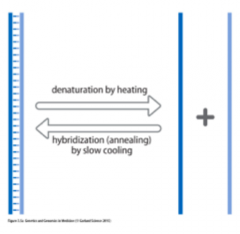
- Amethod for detection ratherthan amplification - Exploitsthe ability of single-stranded nucleic acids to form double-stranded moleculesby complementary base pairing =hydrogen bonding - TwoDNA strands can be separated by heating =Denaturation - Gradualcooling allows separated strands to come together again =Annealing or hybridization |
|
|
Define: Denaturation |
Two DNA strands can be separated by heating |
|
|
Define: Annealing / Hybridization |
Gradual cooling allows separated strands to come together again |
|
|
Define: Artificial Heteroduplexes |
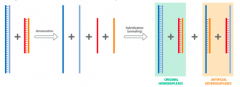
Amixture of two different dsDNA fragments with a high level of sequence identitycan form two types of DNA duplexes: Homoduplexes and Heteroduplexes |
|
|
Define: Homoduplexes |
Perfectbase-pairing over entire length |
|
|
Define: Heteroduplexes |
Base-pairingthat is not perfect across the lengths of the two complementary strands |
|
|
Define: Probe |
Aknown population of nucleic acid molecules or synthetic oligonucleotides |
|
|
Explain: Hybridization Assays |
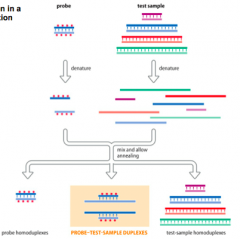
- Probe isused to interrogate a test sample - Eitherthe probe ortest sample must firstbe labeledin some way - Bothare denatured into single strands and then mixed - Theprobe will form artificial heteroduplexes withcomplementary strands in the test sample, if present |
|
|
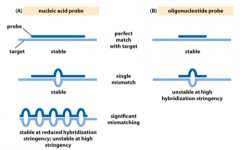
Explain: Hybridization stringency |
- Assayconditions can be adjusted so that only perfect matches are allowed, or so somemismatching is tolerated - Highstringency conditions include high temperature and/or low salt concentration - Bothfactors make hydrogen bonds unstable - Longprobes are more tolerant of mismatching than short oligonucleotide probes - 1 bpmismatch out of 18 bp maycompletely destabilize the duplex, while 1 bp mismatch out of 100 bp willlikely have no effect |
|
|
What are the impacts of Probe length? |
|
|
|
Where is low Hybridization stringency used? |
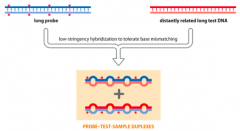
Canbe used to identify related genes (homologs) in the same or different species. |
|
|
Where is high Hybridization stringency used? |
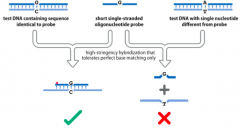
Canbe used to identify alleles that differ by a single base-pair. |
|
|
How are Labelled Probe and Labelled Sample made? |
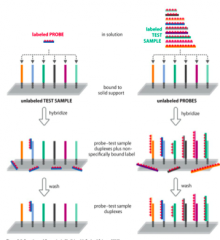
- Inboth approaches the unlabelled population is immobilized on a solidsupport - Hybridizationwith labeled population, followed by wash & detection - Complementarybinding results in immobilization of label |
|
|
Where are Labelled Probes used? |
Labeledprobe is used in Southern & Northern blot, tissue insitu,chromosomein situ |
|
|
Where are Labelled Samples used? |
Labeledsample is primarily used in microarray assays |
|
|
How does PCR Synthesis of DNA Probes look like? |
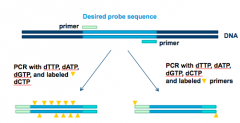
|
|
|
What types of Labelling Systems are there? |

|
|
|
Explain: Microarrays |
- Largescale hybridization to immobilized probes on a high-density grid - Each“feature” (spot)contains millions of identical copies of oneprobe - Fluorescently-labeledDNA or RNA (cDNA) is hybridized to the microarray - Washingremoves nonspecific binding and unbound probe - Microarrayis scanned to detect signal intensity - Reflectsthe number of labeled molecules bound to each feature - Enablesquantitation |
|
|
How do Microarrays work? |
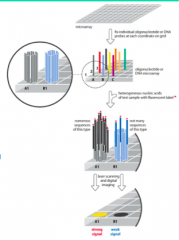
|
|
|
What happens in expression profiling with Two-Colour Microarrays? |
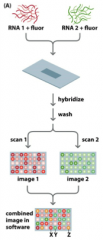
- Two RNAsamples are converted to cDNA samples labeled with different fluorophores • - Hybridizedsimultaneously - Microarrayis scanned to detect signal from eachfluorophore & images are overlayed - Interpretationof signals: - X(red) - Y(green) - Z(yellow) |
|
|
What was the recent predominant method? |
Untilrecently, Sanger Dideoxy Sequencing was the predominant method - Sequencesa single purified DNA fragment - Isstill widely used for investigating particular sequences of interest |
|
|
What new technologies led to the development of massively parallel DNA sequencing? |
Next Generation Sequencing - Sequencesmillions of DNA fragments in a complex sample simultaneously - Allowsfor rapid sequencing of whole genomes |
|
|
What are the Sanger Dideoxy DNA Sequencing components? |
- Asingle oligonucleotide primer - DNApolymerase - UnlabeleddNTPs - Asmall proportion oflabeled ddNTPs - PurifieddsDNA of interest |
|
|
How does Sanger Dideoxy DNA Sequencing work? |

1. TheDNA sequence of interest is first amplified by PCR or cloning, and purified 2. ThedsDNA is denatured, the primer binds to one strand, and polymerase extends fromthe primer, incorporating dNTPs and ddNTPs - Additionof a ddNTPterminates synthesis 3. Typicallyeach ddNTPcarries a different fluorescent label - Randomincorporation of a ddNTPproduces acollection of DNA fragments thathave a common5’ end, butwith variable 3’ ends• - Concentrationof ddNTPs mustbe ~1/100 of dNTPs toensure sufficient extension 4. Fragmentsare separated by size and the fluorescent label on each fragment is detected |
|
|
How does Gel Electrophoresis work? |
1. Nucleicacids migrate through a porous gel in an electric field 2. DNAfragments are separated by size - Agarosegels are used for DNA fragments 100 bp – 20 kb (e.g. PCR products, plasmids) - Polyacrylamidegels have veryhigh resolution ofsmallerDNA fragments (< 1 kb) - Canseparate fragments that differ by a single nucleotide - Usedto separate DNA sequencing products - Slabgel electrophoresis is time & labour intensive 3. The migratingDNA fragments pass a laser that excites the fluorophores and causes emission offluorescence - DNAis visualized with ethidium bromide or Sybr green - Bindto nucleic acids and fluoresce when exposed to UV light - Fluorescentsignals are recorded by a detector |
|
|
Explain: Capillary Gel Electrophoresis |
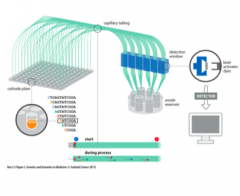
- DNAfragments are separated in long thin capillary tubes containing polyacrylamideor similar gel matrix - 96wells containing DNA fragments are read at once - Increasedautomation |
|
|
Explain: Next Generation Sequencing |
- Thesequencing reaction is monitored, and the DNA sequence is recorded while thestrand is synthesized =Sequencing by synthesis - Manydifferent DNA fragments in a complex starting DNA sample can be simultaneouslysequenced without gel electrophoresis - Somemethods require amplification of the DNA by PCR first, while others useunamplified DNA |
|
|
How does Next Generation Sequencing work? |
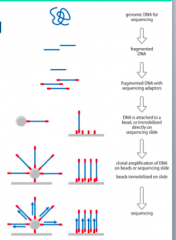
- GenomicDNA is fragmented and adapter oligonucleotides are attached - DNAis attached either to a bead (Roche) or directly to the sequencing slide(Illumina) - DNAis amplified to produce a cluster of molecules with identical sequence - Rochesequences by iterative pyrosequencing - IndividualdNTPs washed over wells one at a time - Lightrecorded when nucleotide is incorporated |
|
|
What are the Iterative Pyrosequencing components? |
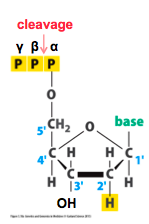
Apyrosequencing reaction includes: - DNApolymerase, ATP sulfurylase,luciferase, and apyrase enzymes - Adenosine5’ phosphosulfate(APS) and luciferin substrates |
|
|
How does IterativePyrosequencing work? |
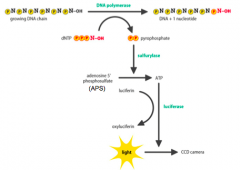
- dNTPincorporation into a growing DNA chain involves cleaving the bond between the α and βphosphates, releasing pyrophosphate (PPi) -Whena dNTP is incorporates: - ATP sulfurylase converts APS and PPitoATP - Luciferase usesATP to convert luciferin to oxyluciferin,which emits LIGHT - dNTPs areprovided individually andin a set order - One dNTP at atime - Lightis only emitted if the provided dNTP is incorporated into the chain - Apyrasedegrades unincorporated dNTPs and any excess ATP, before the next dNTP is added |
|
|
How does Illumina Method work? |
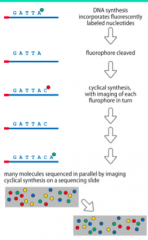
- Alternativesequencing-by-synthesis approach - Usesfluorescently-labeled reversibly terminated nucleotides - Each dNTP hasa terminator + fluorescent label attached to 3’-OH - Asingle nucleotide is incorporated and unbound dNTPs are washed away - Fluorescentsignal is recorded - Terminatorgroup and fluorophore blocking 3’OH are cleaved off & repeat |
|
|
Explain: "Massively Parallel" |
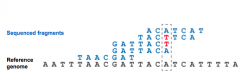
- Eachsequencing slide contains millions to billions of nanowells,each containing a DNA fragment - Allfragments are sequenced simultaneously - Thelength of sequence differs between different methods & sequencers - Rangesfrom ~35 nt –600 nt - Randomfragmentation of genomic DNA results in multiple sequence reads from the sameportion of the genome - Bioinformaticsapproaches align these to the reference genome |
|
|
What are the two key innovations of Single Molecule Real-Time (SMRT) Sequencing? |

- 1) Phospholinked-labeleddNTPs - 2) SMRTChips |
|
|
Explain: SMRT Sequencing (Phospholinked-labeled dNTPs) |
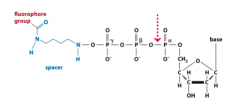
- Each dNTP isattached to a different fluorophore via the γ phosphate - Fluorescencesignal is recorded before cleavage & incorporation - Physicalbulk of fluorophore is released, allowing DNA polymerase to synthesizecontinuously without inhibition |
|
|
Explain: SMRT Sequencing (SMRT Chips) |
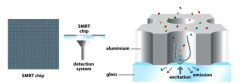
- Containsactive DNA polymerase immobilized at the bottom of a nanowell - Associateswith a single DNA template - DNApol holds dNTP inplace during incorporation - Excitationillumination directed at bottom of chamber causes dNTP toemit a signal identifying the base being incorporated |

