![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
29 Cards in this Set
- Front
- Back
|
mutation categories |
1. somatic mutations: cannot be transmitted to next generation 2. gametic/germ line mutations: passed to half of offspring; all cells of offspring will carry the mutations |
|
|
classification based on phenotype |
1. morphological: chances in appearance 2. nutritional: inability to synthesize essential nutrients 3. biochemical: affect enzyme activity/function 4. behavioral: affect behavior; can have many underlying causes 5. conditional: only noticeable under certain conditions 6. regulatory: involve timing/location of gene expression 7. lethal: cause death |
|
|
types of point mutations |
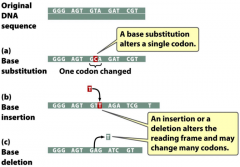
-substitution -insertion -deletion |
|
|
types of base substitutions |
-when a nucleotide pair in DNA is replaaced with a different type of nucleotide base pair 1. transitions: purine to purine (A to G or G to A) or pyrimidine to pyrimidine (T to C or C to T); most common type of substitution 2. transversions: purine for pyrimidine (A or G to C or T) or pyrimidine for purine (C or T to A or G) |
|
|
sickle cell anemia |
-single amino acid change -transversion in the 6th codon: GAG (glutamate) --> GUG (valine) -Hb changes shape --> restricted blood flow --> anemia -but enhanced tolerance to malaria |
|
|
insertions & deletions |
-affect all downstream coding sequences -alteration of protein -truncated due to a stop codon -restoring the reading frame: opposite change occurs; if nearby only a few aa may be altered and can still get a functional protein -large deletions --> loss of genes -many small mutations occur in introns: silent |
|
|
frame shifts |

-results from an insertion or deletions -effects are more severe than substitutions -shift in reading frame |
|
|
types of mutations |
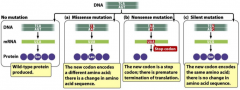
-missense -nonsense -silent |
|
|
phenotypic affects of mutations |
-mutations realized when compared to the wild-type phenotype: forward vs reverse mutation -suppressor mutations: genetic change that hides/suppresses effect of another mutations; different from a reverse mutation in which original mutated site is changed back |
|
|
consequences of mutations |
1. transcriptional level: disruption of TATA box (binding of transcriptional activator protein); mRNA processing (all steps); mRNA stability (protein coding sequence remained intact) 2. translational level: creating a stop codon (UGG tryptophan --> UGA stop); ribosome binding 3. post-translational protein processing: recognition sire for modification may have been changed |
|
|
mutations can result from... |
1. spontaneous mutations: replication errors (mispairing/wobble pairing, strand slippage, unequal corssing over) or chemical changes (depurination, deamination) 2. induced mutations: changes by environmental chemicals or radiation (base analogs, alkylating agents, deamination, hydroxylamine, oxidative reactions, intercalating agents, radiation) |
|
|
wobble pairing |
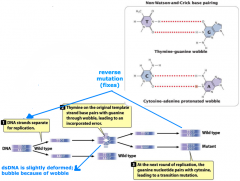
-spontaneous mutation -bubble |
|
|
strand slippage |
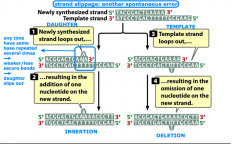
-spontaneous mutation |
|
|
unequal crossing over |
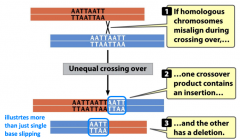
-spontaneous mutation |
|
|
base analong |
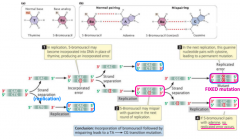
-induced mutation -can substitute for purines or pyrimidines -5-BU: thymine analog that base pairs with G -2-AP: adenine analog that base pairs with C -causes transitions |
|
|
alkylating agents |
-induced mutation -donate alkyl group to nucleotides forming analogs -affect base pairing: transitions -mustard gas, ethylmethane sulfonate (EMS), diethyl sulfate (DES) |
|
|
intercalating agents |
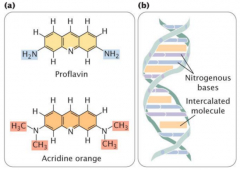
-induced mutation -acridine orange, ethidium bromide, proflavin = intercalating agents that cause frameshift mutations (much more severe) |
|
|
ionizing radiation |
-induced mutation -X-rays, gamma rays, cosmic rays -very short wavelength -high energy radiation ionizes water; forms free radicals (superoxide and hydroxyl) that are highly reactive --> single and double stranded breaks -single = repairable, double leads to chromosome rearrangements -base modification |
|
|
UV radiation |
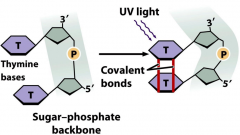
-induced mutation -shorter wavelength = high energy -causes pyrimidine dimers that block replication -lethal -linkage of adjacent pyrimidines on a DNA strand: T-T (common), C-T, C-C -dimers do not normally base pair -stalls replication |
|
|
ABO blood types |
-A and B antigens: sugars found of surface of RBCs -A and B represent modifications of the H substance (carbohydrate group with 3 sugars) -glycosyltransferase modifies H substance -modification based on addition of a terminal sugar: A = N-acetylglucosamine, B = galactose -AB type can add either terminal sugar -O type has no terminal sugar (h substance is only on surface of RBCs) |
|
|
ABO blood types mutations |
-glycosyltransferase mutations -result in A & B modified H substance: 4 nucleotide substitutions differ A & B -type O lacks glycosyltransferase activity: single nucleotide deletion early in coding sequence; frameshift mutation with premature stop codon --> shortened polypeptide chain --> non functional |
|
|
muscular dystrophy |
-muscular degeneration (myopathy) -death due to respiratory failure in early 20s -dystrophin = structural protein in normal muscles -two forms: 1. Becker muscular dystrophy (BMO): less severe, dystrophin detected, mutation doesn't alter the reading frame 2. Duchenne muscular dystrophy (DMD): more severe, more common, no dystrophin detected, mutation alters reading frame |
|
|
trinucleotide repeats |
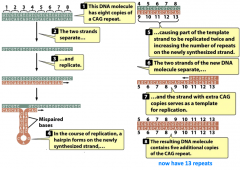
-associated with mutant genes and disease occurrence/severity |
|
|
Ames test |
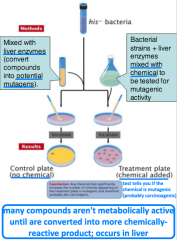
-is a compound mutagenic? -uses 4 strains of Salmonella -specific sensitivities: all are his-, one strain detects base-pair substitutions, three detect frameshift mutations -identifies reverse mutations (his- --> his+) |
|
|
mutation rate |
-expressed as: base pair changes per biological unit (cell division, gamete, generation, etc) -spontaneous mutation rates: low for all organisms, variable between genes, variable between species -bacterial and viral genes: 1/100million cell divisions -mice: 1/10.000 -maize, Drosophila, humans: 1/1.000.000 and 1/100.000 |
|
|
mutation frequency |
-not same thing as mutation rate! -incidence of a specific type of mutation within a group of individuals -mutation frequency of 2x10^-4 means that on average 1/5000 people carries a specific mutation |
|
|
complementation test |
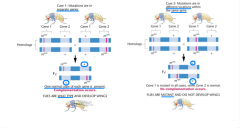
-how can you tell if independent cases are result of same mutation or different mutations? -how can you tell if they are alleles or mutations in separate genes? -how can you tell if two mutations that yield same phenotype present in the same gene or two different genes? --> cross the two mutant strains and analyze the F1 generation: two possibilities |
|
|
penetrance and expressivity |
-gene function or products may not be an absolute y/n -range of phenotypes can be associated with a gene -mutant genotypes: distinct phenotypes, non-distinguishable from normal (wild-type) -penetrance: percent of mutants that express mutant phenotype (e.g. 15% of mutants exhibit wild-type, then mutant gene has a penetrance of 85%) -expressivity: range of expression of the mutant genotype; normal phenotypes to variable mutant phenotypes; background/environmental effects (e.g. normal --> furthers mutation) |
|
|
pleitropy |
-condition where single gene influences more than one trait -doesn't involve multiple functions of a gene; usually related to production of a single substance that is utilized multiple times -production of multiple traits -e.g. inheritance of wrinkled seeds (ww) in peas results in reduced branching of starch in the seeds; plant may also be unable to produce normal starch in other parts like leaves and roots --> one gene but effects are evident in multiple locations |

