![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
107 Cards in this Set
- Front
- Back
|
Order of greatest to least RNA?
|
rRNA>tRNA>mRNA>fRNA
|
|
|
Addition of nucleotide to DNA (draw)
|
Refer to lecture 5.
|
|
|
Promoters
|
DNA sequences that guide RNAP to the beginning of a
gene (transcription initiation site). |
|
|
Terminators
|
DNA sequences that specify the termination of RNA
synthesis and release of RNAP from the DNA. |
|
|
Steps of RNAP
|
1) Initiation.
2) Elongation. 3) Termination. |
|
|
Mapping Promoters
|
DNA sequences that guide
RNAP to the start of a gene (transcription initiation site) |
|
|
Describe the steps of the experiment performed to find where RNAP is on a strand of DNA.
|
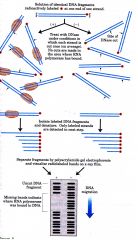
|
|
|
Describe steps to find location of RNAP on DNA strand.
|
1. Radioactively label DNA fragments at one end of strand.
2. Treat with DNAse (No cuts are made where RNAP is bound) 3. Isolate labelled DNA fragments and denature 4. No cuts are made in the area where RNA polymerase was bound. 5. Isolate labelled DNA fragments and denature. Only labelled fragments are detected in the next step. 6. Separate fragments by gel electrophoresis 7. Missing bands indicate where RNA polymerase was bound to DNA |
|
|
Coding Strand is also known as...
|
Nontemplate strand, Sense strand.
|
|
|
Promoters consist of..
|
-35, -10 (Pribnow Box)
|
|
|
The closer the match to the -10 and -35 boxes
consensus... |
the stronger the promoter.
|
|
|
Spacers that are longer or shorter than the
consensus length... |
make weak promoters
|
|
|
Holoenzyme
|
2 α, 1 β, 1 β', 1 σ
|
|
|
Core enzyme
|
2 α, 1 β, 1 β'
|
|
|
Functions of α, β, β', σ?
|
σ recognizes promoter sequences on DNA & is essential for assembly and
for activation of the enzyme; β' binds DNA; β binds NTPs and interacts with σ |
|
|
σ70, σ32, σ60
|
principal σ, heat shock, nitrogen starvation,
|
|
|
σ4 and σ2 bind to which sequences, respectively?
|
-35, -10
|
|
|
An asymmetrical promoter
|
defines which direction the
polymerase will initiate synthesis. |
|
|
Steps of RNAP.
|
1) Binding of RNA polymerase holoenzyme to form a closed complex
2) Formation of an open complex 3) Addition of 6-10 NTPs 4) Dissociation of sigma |
|
|
Trigger loop:
|
A key RNAP loop that recognizes the correct NTP substrate at the
active site of RNA polymerase. It changes its conformation from a disordered loop to two α helices when the correct NTP enters the active site and triggers phosphodiester bond formation. |
|
|
NADFDGD motif
|
To discriminate rNTPs from dNTPs, RNAP recognizes the 2’-
OH group of the ribose via hydrogen bonding with the side chain of an Asn (N) residue found in the universally conserved NADFDGD motif . Mutation of this Asn residue increases dNTP incorporation. |
|
|
Mg+ ions of RNAP
|
-Mg+ ions in active site of RNAP are responsible for phosphodiester bond formation.
-1 of Mg+ ions is responsible for keeping incoming phosphates of nucleotide spatially aligned. |
|
|
If RNAP incorporates a mismatched nucleotide..(write down steps)
|
1) Pause elongation
2) Backtrack to form frayed RNA 3' terminal nucleotide. 3) Gre factor or intrinsic cleavage to remove nucleotides 4) RNA hydrolysis makes a new 3' OH group at end of RNA to restart elongation. |
|
|
Rho Dependent Termination
|
Rho factor binds the end of RNA chain and slides along the strang towards the open bubble complex. When the factor catches polymerase, it causes termination of transcription.
|
|
|
Rho Independent Termination
|
You should be able to draw stem loop structure first!
-Because the uracils will only weakly pair with the adenines, the RNA chain can easily be released from the DNA template, terminating transcription. |
|
|
Translocation
|
Once the correct nucleotide is added, the TEMPLATE has to be moved such that the next nucleotide is positioned correctly in the active site
of RNAP New data suggest a role for key structural elements of RNAP in this process |
|
|
Rho
|
ATP-dependent helicase
– it moves along the RNA transcript, finds the "bubble", unwinds it and releases the RNA chain. |
|
|
How does RNAP let go of the promoter and translocate downstream?
|
Synthesis of RNA chains destabilize the interactions between Sigma 4 and the Beta flap (destabilizing interaction between the -10 and -35 elements), which allow RNAP to let go of the promoter.
|
|
|
Rut Site
|
Rho initially binds to rut site on mRNA
|
|
|
Allosteric Model of Rho Termination
|
Termination induced by nascent RNA that alters the RNAP elongation complex
|
|
|
Positive Regulation (activation)
|
a positive regulatory
factor (activator) improves the ability of RNAP to bind to and initiate transcription at a weak promoter. |
|
|
Negative Regulation (repression)
|
a negative regulatory
factor (repressor) blocks the ability of RNAP to bind to and initiate transcription at a strong promoter |
|
|
polycistronic mRNA
|
An mRNA that codes for more than one protein.
|
|
|
Operons
|
A unit made up of linked genes that is thought to regulate other genes responsible for protein synthesis.
|
|
|
Operators / Enhancers
|
-Operators = Repressor Binding Sites
-Enhancers = Activator Binding Sites |
|
|
Constitutive genes
|
are expressed all of the
time. i.e. glucose metabolizing |
|
|
Inducible / Adaptive genes
|
are normally off and are
turned on by the presence of an inducer substance. i.e. lactose metabolizing |
|
|
Repressible genes
|
are normally on and are
turned off by the presence of a repressor substance. |
|
|
repressor protein (LacI)
|
It is an allosteric protein
with two binding sites: either binds Operator DNA or inducer (allolactose). |
|
|
The lac promoter region has two binding sites:
|
1) RNA polymerase binding site
2) CAP-binding site (for catabolite activator protein), an example of positive control |
|
|
What activators work by Allostery rather than Recruitment of RNA polymerase?
|
NtrC, MerR
|
|
|
NtrC
|
interacts with its DNA binding site only after phosphorylation by NtrB in response to nitrogen starvation MerR
|
|
|
MerR
|
Binds DNA, but Alters its Conformation only in Response to Mercury Binding
|
|
|
NusA
|
-elongation factor, promotes termination
-NusA enables the formation of stable ECs (elongation complexes), which leads to processive transcription and read-through of termination sites. |
|
|
NusG
|
-is an essential bacterial regulator of the RNAP
-enables transcription:translation coupling, or to ρ factor, to support ρ-dependent termination. |
|
|
NusB
|
-has single stranded RNA binding activity and forms a stable complex with NusE. This
heterodimer formation enhances the RNA binding affinity of the complex. |
|
|
NusE
|
NusE is involved in translation
|
|
|
Eukaryotic Promoter
|
TATA box (analagous to -10 box), CAAT box (-30 in eukaryotes), GC box.
|
|
|
Core promoter elements? Proximal promoter elements?
|
Core promoter elements = TATA box; Proximal promoter elements = CAAT + GC box.
|
|
|
Deletion of TATA box
|
heterogeneity of transcript start
site without eliminating transcription |
|
|
Distant-independent elements
|
silencers / enhancers
|
|
|
Transcription Factors
|
Trans acting factors that alter
the rate of transcription. Often bind to DNA. |
|
|
Transcriptional activators
|
increase rate of transcription
|
|
|
Transcriptional repressors
|
decrease the rate of
transcription |
|
|
Common Domains of transcription Factors
|
1) DNA binding domain [binds to DNA sequences]
2) Activation domain [Interacts with basal machinery to elevate the rate of transcription] 3) Dimerization domain [mediates protein-protein interaction.] 4) Ligand binding domain [the function of some transcription factors is altered by the binding of a molecule] |
|
|
Basal Transcription Factors
|
-factors required for initiation of
transcription of genes; bind to a core promoter (TATA box, -30) |
|
|
Basal Transcription Factors for RNA Polymerase II
|
TFIID, TFIIA, TFIIB, RNA Polymerase II, TFIIF, TFIIE, TFIIH.
|
|
|
TFIID
|
-Consists of TBP (TATA box binding protein) + TAFs (TBP associated factors).
-Bind to core promoter -Kinks DNA at each end of the TATA box -First step in basal transcription is binding of TFIID to core promoter -Other core factors bind to TFIID |
|
|
TFIIA
|
Increase affinity of TBP for DNA in vitro.
|
|
|
What are the core factors? (Proteins that bind the promoter)
|
TFIIA, TFIIB
|
|
|
Preinitiaton Site (PIC)
|
a large complex of proteins that is necessary for the transcription of protein-coding genes in eukaryotes (+archaea). The preinitiation complex helps position RNA polymerase II over gene transcription start sites, denatures the DNA, and positions the DNA in the RNA polymerase II active site for transcription
|
|
|
TFIIH
|
has both helicase
and kinase activities that can unwind DNA and phosphorylate the CTD tail of RNA Pol II |
|
|
YS2PTS5PS7
|
CTD heptapeptide repeats of RNA Polymerase II.
-Phosphorylation of these serine repeats during promoter escape / capping / splicing / poly A addition. |
|
|
How does addition of the polyA tail occur?
|
-RNA polymerase II transcribes well past the end of the gene
-After the end of the gene has been reached, RNA polymerase II passes through an AATAA sequence, which lies beyond the 3' end of the coding sequence. 3. The pre-mRNA, carrying this signal, AAUAA, is then cleaved by a special endonuclease that recognizes the signal and cuts at a site 11 to 30 residues to its 3' side. |
|
|
Poly A Polymerase (PAP)
|
-Poly A polymerase adds A's to the 3' end.
-Binding of poly A binding protein (PABP) enhances the rate of poly a polymerase |
|
|
Why is capping /polyadenylation of mRNA at the 3' end important?
|
-Transport of DNA
-Protects from nucleases -Stabilizes |
|
|
Rat1 Protein
|
-Protein present on RNA pol II
-Loads onto remainder of transcript and starts chewing 5' --> 3' direction. -Promotes termination when reaches RNAP , allosterically altering it. |
|
|
Alpha Amantin
|
From the amanita mushroom
|
|
|
Actinomysin D
|
Inhibits all RNA polymerases; blocks elongation
|
|
|
Rifampicin
|
Blocks promoter clearance / inhibits elongation
|
|
|
mRNA processing in Prokaryotes vs. Eukaryotes
|
-Prokaryotes: no significant modification
-Eukaryotes: a) 5' capping b) 3' cleavage and polyadenylation c) removal of introns (nuclear splicing) |
|
|
C Terminal Domain of RNA Polymerase
|
Has serine repeats that are differentially modified as the cell goes through different stages (that recruit different processing factors)
|
|
|
Forward Translocation
|
Forward movement about RNAP with respect to RNA/DNA hybrid by 1 or several nucleotides without transcript elongation, thus promoting release of transcript.
|
|
|
Roles of Nus G / Nus A in termination
|
Nus G = Rho dependent termination
Nus A = Rho independent termination by helping to form the stem loop structure. |
|
|
IPTG/XGAL
|
Allolactose analogs
|
|
|
Pribnow Box
|
A/T rich, ideal for unwinding
|
|
|
Sanger method (Chain termination)
|
-ddNTPs will be radioactively or fluorescently labelled
-The DNA sample is divided into four separate sequencing reactions, containing all four of the standard deoxynucleotides (dATP, dGTP, dCTP and dTTP) and the DNA polymerase. To each reaction is added only one of the four dideoxynucleotides (ddATP, ddGTP, ddCTP, or ddTTP) which are the chain-terminating nucleotides, lacking a 3'-OH group required for the formation of a phosphodiester bond between two nucleotides, thus terminating DNA strand extension and resulting in DNA fragments of varying length. -newly synthesized and labelled DNA fragments are heat denatured, and separated by size by gel electrophoresis -the DNA bands are then visualized by autoradiography or UV light, and the DNA sequence can be directly read off the X-ray film or gel image -A dark band in a lane indicates a DNA fragment that is the result of chain termination after incorporation of a dideoxynucleotide -The relative positions can then be used to read a DNA sequence |
|
|
Maxam-Gilbert method of Sequencing
|
-The method requires radioactive labeling at one 5' end of the DNA and purification of the DNA fragment to be sequenced.
-End-label with radioactive phosphate and chemically cleave |
|
|
How do you weed out recombinant λ phage vectors?
|
λ phage vectors that have failed to acquire a foreign DNA insert are unable to propagate because they are too short to form infectious phage particles.
|
|
|
LacZ gene of the pUC18 plasmid cleaves what? What color is the product?
|
-lacZ gene in the pUC18 plasmid (see Fig. 3-24) encodes the enzyme β-galactosidase, which cleaves the colorless compound X-gal to a blue product
|
|
|
How do you weed out recombinant vectors using the LacZ gene of the pUC18 plasmid?
|
-if the plasmid contains a foreign DNA insert in its polylinker region, the colonies are colorless because the insert interrupts the protein-coding sequence of the lacZ gene and no functional β-galactosidase is produced.
-If the ligation was successful, the bacterial colony will be white; if not, the colony will be blue. |
|
|
How is a cDNA library made?
|
cDNA library is constructed by isolating all the cell's mRNAs and then copying them to DNA using a specialized type of DNA polymerase known as reverse transcriptase because it synthesizes DNA using RNA templates (Box 25-2). The complementary DNA (cDNA) molecules are then inserted into cloning vectors to form a cDNA library.
|
|
|
Back-tracking
|
The process of RNA polymerase stopping forward movement and backing up by
a few bases, ~about five bases |
|
|
Anti-termination
|
A process whereby RNAP is converted to a form that is more resistant to
termination. |
|
|
β' trigger–loop
|
RNAP loop that recognizes the correct rNTP substrates at the active site of RNA polymerase. It changes its conformation from a disordered loop to two α
helices when the correct rNTP enters the active site and triggers phosphodiester bond formation. |
|
|
NADFDGD
|
The absolutely conserved NADFDGD motif, present in all RNA polymerases, that
is involved in coordination the active site Mg2+ ions, which are essential for catalysis |
|
|
heptapeptide
|
The YSPTSPS conserved sequence many copies of which are found on the Cterminal
domain of RNAP II. The conserved ser residues are differentially phosphorylated at various stages of transcription. |
|
|
Footprinting
|
The process of determining the binding site of a protein or enzyme (RNA Polymerase) on DNA.
|
|
|
Genes required for growth under oxidative stress
|
Sigma^oxstress
|
|
|
Genes required for growth under cold shock conditions.
|
Sigma^coldshock
|
|
|
Genes required for stationary phase growth
|
Sigma^S
|
|
|
Genes required for growth under high pressure
|
Sigma^HiPressure
|
|
|
ORF
|
is a DNA sequence that does not contain a stop codon in a given reading frame
|
|
|
Open Complex
|
the complex of RNA polymerase and one DNA strand (The antisense strand)
|
|
|
Closed Complex
|
complex of RNA Polymerase when it is bound to the double stranded DNA
|
|
|
Silencer
|
Silencers are control regions of DNA that, like enhancers, may be located thousands of base pairs away from the gene they control. However, when transcription factors bind to them, expression of the gene they control is repressed.
|
|
|
Activator
|
DNA-binding protein that regulates one or more genes by increasing the rate of transcription. The activator may increase transcription by virtue of a connected domain which assists in the formation of the RNA polymerase holoenzyme, or may operate through a coactivator.
|
|
|
Coactivator
|
A coactivator is a protein that increases gene expression by binding to an activator (or transcription factor) which contains a DNA binding domain.
|
|
|
Proximal elements
|
the proximal sequence upstream of the gene that tends to contain primary regulatory elements
|
|
|
Distal Elements
|
the distal sequence upstream of the gene that may contain additional regulatory elements, often with a weaker influence than the proximal promoter
|
|
|
Consensus Sequence
|
a sequence of nucleotides or amino acids similar or identical between regions of homology in different but related DNA, RNA, or protein sequence.
|
|
|
AAUAA
|
Sequence on the 3' end that signals cleavage of mRNA by poly A polymerase some 20 nucleotides downstream of the signal.
|
|
|
Downstream
|
3' End
|
|
|
mRNA cap
|
5' End
|
|
|
How do restriction enzymes protect themselves?
|
EcoRI methylase adds a single methyl group to the recognition sequence.
|
|
|
DNA Ligase
|
carries out ligation by using ATP energy to make the phosphodiester bond between the vector and passenger.
|
|
|
On the other hand, cDNA libraries (c stands for copy) are made by first converting mRNA into a DNA copy, a process known as____
|
Reverse transcription
|

