![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
94 Cards in this Set
- Front
- Back
|
Micrococcal Nuclease Digestion
|
Endo-exonuclease. Preferentially digests at A an or T than G or C. Leaves mono, di, tri- nucleosomal ladders
|
|
|
Requirements for nucleosomal arrays
|
DNA, ATP, histone chaperone (NAP1), chromatin remodeler (CHD1)
|
|
|
Replication- required proteins
|
MCM- helicase
PCNA- sliding clamp, ensuring processivity of DNA POL Two different DNA Pols, leading and lagging strand |
|
|
How many types of histone chaperones?
|
Four; two for H2A/H2B and two for H3/H4
|
|
|
CAF1
|
H3/H4 tetramer import/assembly. Required for DNA replication, three subunits: first (Cac1) binds PCNA, 2nd (Cac2) binds ASF1, 3rd (Cac3) binds histones, deposits histones after replication fork. Heterchromatin maintenance
|
|
|
Histone synthesis occurs
|
In G1/S phase
|
|
|
How many base pairs before and after the replication fork are naked?
|
250 bp
|
|
|
Histone Chaperone Functions/Characteristics
|
Negatively Charged
Help in Chromatin Assembly Prevent Aggregation of Histones |
|
|
ASF1
|
H3/H4 dimer assembly, then brings to CAF1 (interacts with Cac2 subunit). Binds to the dimerization region of H3/H4 dimers, so does not do tetramers. Cannot assembly on its own. Disassembly- linked to MCM and import. Can bind newly synthesized and parental histones.
|
|
|
NAP1
|
H2A/H2B (H3/H4) IMPORT, assembly, disassembly. Can assemble chromatin in vitro (regular spacing with CHD1)
|
|
|
FACT
|
H2A/H2B (H3/H4). Assembly, disassembly. Interacts with DNA POL and MCM and localizes to the replication origin. Promotes replication initiation
|
|
|
What part of the nucleosome does H2A and H2B contact?
|
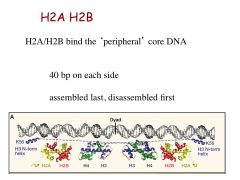
The peripheral core DNA, 40 bp on either side, assembled last, disassembled first
|
|
|
Histone deposition: Modifications
|
Newly synthesized histones have acetylated tails. Acetylated H3 and H4 tails are different from other acetylation and required for assembly.
Done by cytoplasmic histone acetyltransferase: HAT1 H3K56ac (yeast) H4K5/K14ac |
|
|
Histone deposition: H4 tail acetylation
|
Nuclear import, CAF1 interaction
|
|
|
Histone deposition: H3K56ac
|
In histone core, loosens DNA interaction, nucleosome assembly, CAF1 interaction- stronger with the H3K56ac
|
|
|
Replication: ATP-dependent Chromatin Remodelers
|
ISWI (nuclesomal arrays, periodicity, heterochromaid replication
CHD1(Nucleosomal arrays) -long arrays and properly spaced nucleosomes require these |
|
|
ISWI in replication
|
Chromatin Assembly, with NAP1 linker length formation of nucleosomal arrays. Transcription elongation, silencing, chromosome organization.
-a tail that it uses as a molecular ruler to measure distance to next nucleosome |
|
|
CHD1: replication
|
1. Chromatin assembly
2. Together with NAP1 3. Histone variant assembly |
|
|
How are epigenetic marks copied after replication?
|
1. random segregation of new and parental histones on new and parental strand. (or combination of both H3/H4 and H2A/H2B)
2. Some mods are regenerated by copying from parental histones- usually by an enzyme/complex that has reader and writer for the same modification 3. Some regulators remain associated with the chromatin through replication 4. Temporal regulation for different types of chromatin |
|
|
Timing of heterochromatin
|
Late replication. Factors required for heterochromatin formation are at the replication fork at this time
DNA methylation, H3K9me3, presence of HP1 |
|
|
Histone Occupancy
|
# of nucleosomes in a given spot
high= one spot is always occupied by a nucleosome |
|
|
Histone Positioning
|
Concordance of nucleosomes in a given region. High= exactly the same spot. Low = variable spots, wider band on a gel/sequencing
|
|
|
How do you make a heat map of nucleosome positioning?
|
MNase digestion, align sequence based on TSS. See +1 and -1 nucleosome enrichement around TSS. Before TSS= Nucleosome free region/nucleosome depleted region
|
|
|
Nucleosome positioning across different genomic regions
|
unstructured on non-genes, -1, nucleosome free region (GTRNX machinery), TSS, +1, +2, structured over genes
|
|
|
What generates the positioning?
|
a. Stretch of A and T= very unbendable, unfavorable for nucleosome position (in yeast), also nucleosome free regions in the terminators
b. Mammals, less well understood, maybe CpG islands?? c. Periodicity of AA, TT, TA every 10 bp in the major groove of the DNA, and have CG in the minor groove nicely ideally bends the DNA d. However, DNA is a poor predictor, 50% or less (same as by chance prediction) e. other proteins could be contributing: TF or POl II or well positioned (container) nucleosome |
|
|
Chromatin Assembly during Transcription
|
Replication independent, linked to incorporation of histone variants. Activities track with elongating Pol II. Genes: consequence of disassembly during transcription. Promotors: nucleosome evition or exchange. Major assembly in post-mitotic cells such as neurons
|
|
|
HirA
|
Replication-Independent Chaperone H3.3/H4 dimer- histone variant during transcription
|
|
|
SWR
|
H2A.Z/H2B deposition during transciption
|
|
|
DAXX
|
H3.3/H4 @ telomeres
|
|
|
Chz1
|
H2A.Z/H2B
|
|
|
Epigenetic Phenomena
|
heritable alternative states of gene activity that do not result from altered nucleotide sequence
|
|
|
Examples of Epigenetic Phenomena
|
arthritis in monozygotic twins, environmental component in disease etiology. 3 year old twins more similar epigenomes than 50 yr old twins. Calico coat color cannot be cloned, due to random X-inactivation= epigenetic
|
|
|
Differences between genetic and epigenetic
|
Genetic: mutations, inherited, germ line, invariable
Epigenetic: Alterations, stable?, soma + germline, variability |
|
|
Heritable vs Not Heritable
|
A transient change that lasts less than one cell cycle= not heritable
A change that can be inherited mitotically or meiotically = heritable |
|
|
Meiotically Heritable Example
|
Toadflax symmetry--> determined by DNA methylation. Inherited through generations
|
|
|
Vinclozolin and Rats
|
Exposure to vinclozolin, an fungicide and anti-androgenic compound, reduce sperm counts in subsequent generations
|
|
|
Sweeden food shortage
|
Grandchildren had increased lifespan if grandpa had less food. Shorter lifespan due to heart disease and diabetes were associated with access to food for grandpa
|
|
|
How compact does the DNA need to be to fit in the nucleus?
|
10,000X
DNA= 2 m long Nucleus is 10 uM radius, accomplished by DNA compaction with proteins. Template for epigenetic phenomena |
|
|
Beads on a string
|
10 nm fiber- histones plus DNA
|
|
|
Secondary Chromatin Structure
|
30 nM fiber- has architectural proteins such as linker histone (H1), heterochromatin protein (HP1), Polycomb group proteins (PcG). One start helix vs two start helix models--> both occur in vivo and depend on the linker length
|
|
|
Tertiary Chromatin Structure
|
Higher order Chromatin Fiber
|
|
|
Nucleosome definition
|
Histone octamer plus 147 bp of DNA, wrapped 1.7 times
|
|
|
Core vs Linker DNA
|
Core= 147 bp wrapped around the histone octamer
Linker= DNA between the histone octamer |
|
|
Histone Conservation
|
H2A- conserved
H2B- conserved H3- highly conserved H4- very highly conserved linker/H1 - not conserved |
|
|
Three histone domains
|
1. Histone Fold- 3 conserved alpha helices
2. Histone Fold Extension- 3. Extended N- and C-termini |
|
|
Description of Histone Proteins
|
Small, 10 kDa, very basic
|
|
|
Description of Histone Tails
|
Lysine and Arginine rich (K, R)
|
|
|
Nucleosomes and transcription
|
Nucleosomes are a block to RNA Pol II, as is higher order packing of chromatin!
|
|
|
Linker DNA
|
Linker length can vary from 10-50 bp. Regular linker length found in inactive chromatin. Irregular linker length found in active chromatin.
|
|
|
Euchromatin
|
Transcriptionally active, less compacted
Gene rich |
|
|
Heterochromatin
|
Less transcriptionally active, very compacted
Gene poor a) constitutive heterochromatin centromeres, telomeres b) facultative heterochromatin rDNA, transposons, inactive X chromosome (Barr Body) |
|
|
Mechanism exist to “open up” chromatin
|
Chromatin remodeling complexes
alter primary structure of chromatin Histone modifying enzymes alter histone tail modifications |
|
|
Mechanism exist to “condense” chromatin
|
Histone modifying enzymes
alter histone tail modifications DNA methylases, Recruitment of chromatin binding proteins Polycomb proteins Heterochromatin Protein |
|
|
Chromatin Regulation vs Epigenetic Regulation
|
Non-heritable chromatin state vs heritable chromatin state
|
|
|
What family do the ATP-chromatin remodelers come from?
|
The SF2 family of helicases, but no helicase activity
|
|
|
What are the four most well-studied classes of ATP-chromatin remodelers?
|
CHD, Ino80/SWR1, ISWI, Swi2/Snf2
|
|
|
ATPase Domain
|
All remodelers share this domain: DExx and HELICc
|
|
|
SWI/SNF domain is...
|
Bromodomain: binds to acetylated lysines
|
|
|
ISWI domain is the ....
|
SANT - SLIDE domain: interacts with N termini of histones
|
|
|
CHD domain is the ...
|
Tandem chromodomains: binds methylated lysines on histone tails
|
|
|
INO80 family domain is ....
|
Long insertion between the DExx and HELICc ATPase domains
|
|
|
Six functions of ATP chromatin remodelers
|
1. Deposition - assembly
2. Repositioning - site exposure 3. Ejection- site exposure 4. Unwrapping- site exposure 5. Dimer exchange - Altered Composition 6. Dimer ejection- Altered Composition |
|
|
Shared Characteristics of ATP remodelers
|
bind nucleosomes
are DNA-dependent ATPases recognize histone modifications ATPase activity can be regulated interact with other proteins |
|
|
How many remodelers per nucleosome?
|
one. Two if you are ISWI
|
|
|
How is the nucleosome destabilized?
|
Multiple enzymatic reactions
|
|
|
How many DNA contacts must be broken and how much energy per contact?
|
14 DNA contacts, 1 kcal/mol/contact
|
|
|
Where does the energy come from for chromatin remodelers?
|
ATP hydrolysis
|
|
|
Where does the chromatin remodeler bind the nucleosome?
|
Near they dyad- two helical turns
|
|
|
Which direction and how fast is the translocation?
|
3' to 5' but can reverse direction, 25 bp per second
|
|
|
How does the remodeler work physically with the DNA?
|
generate DNA loops, reposition nucleosome or destabilize
|
|
|
Restriction Enzyme Accessibility Assay
|
• Template has various RE sites in either linker or nucleosomal sites
B is freely accessible, A is not once a remodeler added, site A now accessible, B is not Look at rate of digestion in presence or absence of remodeler Very similar to MNase digestion assay |
|
|
Octamer Transfer
|
• Octomer on DNA not radioactive, then add RA DNA, and see if the remoderler puts octamer on RA
• Free DNA migrating very rapidly, then after, get a fraction that migrates more slowly which means the nucleosome has be remodeled |
|
|
o Altered topology on a closed circular array
|
• Circular plasmid assembled with nucleosomes
• when you add chromatin remodelers, you can add more octamers to the plasmid • Now entire plasmid is assembled with octamers • if you treat with topoisomerase and deprotonize it- proteinase K, get it relaxed, and migrate slowly, more octamers, concatemated DNA and migrates rapidly -Output is the activity of the remodelers -Ladder is proportional to the number of octamers added |
|
|
Altered DNAase digestion
|
more cleavage sites with DNAase + remodeler
|
|
|
o MNase/chemical method from sam’s paper, followed by qPCR, seq, or H3 ChIP seq (histone H3 ChiP)
|
• Labor intensive, indirect, and hard, but important
|
|
|
ISWI / SNF2H- size and number of complexes
|
small complexes, MANY different ones
|
|
|
ISWI / SNF2H- Roles
|
o nucleosomal arrays assembly
o important for H-chromatin formation o reprogramming (nuclear transfer) o Transcriptional regulation some Pol III, PolI o Famous for positioning unfavorable positions on DNA • This kind of activity is important at the end of genes to keep promiscuous transcription from initiating o Slide 22- stain for Iswi, many loci of genome have presence of both factors, doesn’t mean they are actually overlapping o 23- inactive X chromosome in the Male X, not properly conserved in the Iswi mutant, no longer properly condensed as they should be |
|
|
ISWI Hand-Sant-Slide Domain
|
binds linker (30 bp from nucleosome)
Mutations in vivo: role in rate of ATP hydrolysis and remodeling |
|
|
Role of ISWI domains
|
SANT/HAND domain contacts histone tails
- charge: histone tail interaction + charge: DNA interaction Slide domain linker DNA contact, ’measures’ distance equal spacing of nucleosomes- first 30 bp ATPase domain near dyad, motor, translocation |
|
|
How does ISWI accomplish equal nucleosomal spacing?
|
Two molecules in opposite directions, one pulls first, then the other pulls back to get even spacing
|
|
|
CHD subgroup members
|
CHD1: role in chromatin assembly; open chromatin
in pluripotent cells CHD3, 4 HDAC complex subunits! NuRD complex also contains Me-DNA binding protein (MBD2) complex connects deacetylation, chromatin remodeling and DNA methylation; repressive function CHD7: together with PBAF; CHARGE syndrome neuronal development Together with SNF2: role in transcriptional elongation |
|
|
Role of CHD chromodomains
|
Regulate/gate the ATPase domain- when deleted, ATP hydrolysis rate much faster
|
|
|
More similarities between CHD1 and ISWI
-mechanism |
Have auto-inhibitory domains (basic residues)
Activity stimulate by unmodified H4 tail (inhibited when acetylated) Helps displace auto-inhibitory domains Of note additional accessory subunits in complexes may further modulate the actions of these ATPases to execute diverse nuclear functions |
|
|
CHD1 and ISWI- chromatin assembly
|
ATP-dependent chromatin assembly is functionally distinct from remodeling
|
|
|
More similarities between ISWI and CHD1
|
1. Play a role in chromatin assembly
2. Act on coding regions of genes (roles in transcription) 3. Role in regular spacing of nucleosomes |
|
|
Roles of CHD chromatin remdolers in vivo?
|
1. Are frequently mutated in endometrial and prostate cancer
2. CHD7 acts together with SWI/SNF ATPase BRG1 in neuronal development 3. CHD4 Thymocyte development 4. CHD1 activation of development (zygote) |
|
|
SWI2/SNF2 Family- Size of complexes, number of members
|
HUGE!
• 30 members of swisnif members in humans, 17 in yeast, 42 in arabadopsis |
|
|
ROLE of SNF2/BRM domains
|
BROMO domain
binds acetylated lysines on histone tails (H3/H4) HSA domain protein interactions actin/ARP transcription factors ATPase domain near dyad, motor, translocation SNAC domain- required for remodeling activity |
|
|
SNF2 ATPase activities-
|
1. Change nucleosome position to increase access for other regulatory proteins
2. Change nucleosome conformation 3. Eject histone octamer 4. Displace H2A/H2B dimer |
|
|
Roles of SWI2/SNF2
|
1. Inducible gene expression- trnx initiation, trnx elongation
2. Splicing 3. Repair 4. Roles in development and stress processes 5. Can be activating or repressive! 6. Only 6% of genes are dependent on SWI/SNF for gene activation 7. Persistence- slide 16 8. Can dock on the entire nucleosome |
|
|
In Vivo roles of SWI/SNF
|
Mammals:
pluripotency (ESC) determination of cardiogenic potential neuronal differentiation and cognition stress response and longevity (C.elegans, Riedel Nature Cell Biol 2013) potent tumor suppressors (mutated in 19% of human cancers (p53 is 26%)) |
|
|
INO80 roles
|
chromatin assembly/replication block- helps unstall the fork
DNA repair interacts with phosphorylated H2A.X (gammaH2A.X) transcription exchange H2A.Z with H2A at -1 and +1 nucleosomes-->replication independent effect |
|
|
SWR1 roles
|
H2A exchange with H2A.Z
Boundary to heterochromatin spreading transcriptionally poised promoters (together with H3.3) |
|
|
Chromatin Remodeler Regulation
|
1. Recruitment by TFs
2. Chromatin environment, histone tail modification 3. Post-translational modifications phosphorylation, acetylation, de-ubiquitylation 4. Complex composition regulatory subunits (SWI5, Drososphila) tissue specific subunits (BAF60c) 5. Allosteric regulation by specific domains (CHD1, ISWI) 6. Interaction with small molecules (phosphatidyl inositol) |

