![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
59 Cards in this Set
- Front
- Back
|
Name the 3 parts of a nucleotide.
|
-deoxyribose sugar
-one phosphate group attached to the sugar -a nucleoside or base: A, T, C, and G |
|
|
What type of bonds link nucleotides or ribonucleotides?
|
covalent bonds between sugar and phosphate
|
|
|
What type of bonds link the bases to create a double helix?
|
hydrogen bonds
|
|
|
Name characteristics of DNA.
|
-Long, double-stranded molecules of DNA.
-Always unbranched -Same four monomers: A, T, C, and G. |
|
|
What is required of the template strand?
|
Replication requires a template strand of DNA. Strand must be free or unpolymerized (hydrogens broken and bases exposed)
|
|
|
What is a semiconservative replication?
|
Each DNA strand is used to synthesize a new double stranded molecule of which it makes up one-half.
|
|
|
What direction does DNA polymerase read a strand?
|
5' to 3'
|
|
|
What is the leading strand?
|
the strand that is continuously polymerized in the 5' to 3' direction
|
|
|
What is semiconservative replication?
|
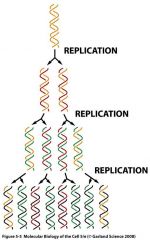
|
|
|
What does a strand of DNA look like at the molecular level?
|
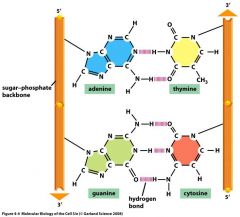
|
|
|
What does it mean that Double stranded DNA runs antiparallel?
|
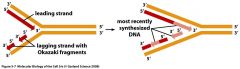
one side of the replication fork is moving in s 3' to 5' direction at all times.
|
|
|
Can DNA synthesis begin de novo?
|
Nothe DNA polymerase requires a primer to begin synthesis.
RNA synthesis, however, can begin de novo intermediate RNA molecules serve as DNA synthesis primers. |
|
|
What types of proof reading methods are used when replicating DNA?
|
-Several methods of proofreading occur from the DNA polymerase itself.
-The correct nucleotide has higher affinity for the polymerase because the reaction is energetically favorable. -When the nucleotide is bound to the DNA polymerase, the enzyme undergoes a conformational change in order to covalently link it to the previous nucleotide. |
|
|
How many primers are required for the lagging strand?
|
For the lagging strand, one primer is required for every Okazaki fragment
|
|
|
What is required to initiate synthesis of DNA?
|
an RNA primer
|
|
|
How many primers are required for the leading strand?
|
One
|
|
|
What is DNA primase?
|
DNA primase
synthesizes short (10bp) RNA primers on DNA templates Results in a DNA:RNA hydrogen-bound hybrid molecule. |
|
|
What does DNA polymerase do after DNA primase performs its actions?
|
DNA polymerase elongates the RNA primer from the 3’-OH end.
|
|
|
DNA primase and polymerase
|
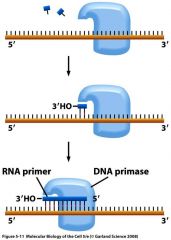
|
|
|
What happens when DNA reaches the 5' end of a primer?
|
It falls off
Creates a hanging single-stranded ribonucleoside which is then Recognized by specialized proteins which remove the RNA primer. DNA ligase then extends the DNA strand from the 3’ end of the new DNA fragment to the 5’ end of the latest DNA fragment. |
|
|
Once RNA primer is removed what does DNA ligase do?
|
extends the DNA strand from the 3' end of the new DNA fragment to the 5' end of the latest DNA fragment essentially gluing the 2 pieces together.
|
|
|
The DNA polymerase and DNA ligase in action
|
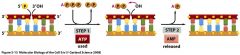
|
|
|
Name proofreading mechanisms.
|
-Incorrect nucleotide addition
At the end of a primer strand will result in that nucleotide remaining unpaired. -There is a separate catalytic site on the DNA polymerase, responsible for exonucleolytic proofreading -It will recognize this unpaired 3’-OH end as incorrect and excise the unpaired nucleotides until a correctly base-paired 3’-OH remains. |
|
|
At what end is it possible to correct mistake in nucleotide addition?
|
3’-OH end.
|
|
|
What isn't it possible to excise or correct a mistake in nucleotide addition from the 5' end?
|
The 5' end would result in a nucleoside monophosphate at the terminus
No further reaction could proceed because the cleavage of a high energy nucleoside triphosphate bond is required for polymerization. |
|
|
What is DNA helicase?
|
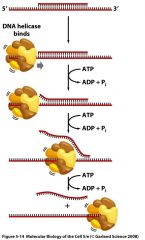
-an enzyme which rapidly moves along a single strand of DNA ahead of the replication machinery
utilizes ATP hydrolysis to break hydrogen bonds between annealed DNA strands thereby opening the replication fork. |
|
|
What happens after helicase has uncovered a DNA strand?
|
Single-strand binding proteins (SSB) bind to the DNA and
aid the helicase in stabilizing the unwound DNA without covering the bases enough to impede templating. |
|
|
What is the purpose of a sliding clamp and where does it bind?
|
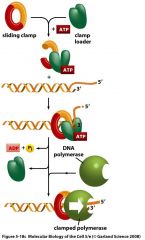
-binds to the back of the DNA polymerase and forms a ring around the DNA molecule.
-prevents the DNA polymerase from frequently falling off -clamp is assembled on the DNA before the polymerase binds it via a clamp loader which requires ATP. -There is one pre-loaded clamp for every RNA primer. |
|
|
What is a sliding clamp?
|
A sliding clamp
Binds the back of the DNA polymerase and forms a ring around the DNA molecule. |
|
|
Where does a sliding clamp bind and when does it bind?
|
Binds to the black of the DNA polymerase and forms a ring around the DNA molecule.
The clamp is assembled on the DNA before the polymerase binds it via a clamp loader |
|
|
What is the purpose of a sliding clamp?
|
preventsthe DNA from frequently fall off
|
|
|
Does a sliding clamp use energy to bind?
|
Yes, ATP
|
|
|
How many clamps does each RNA primer use?
|
one
|
|
|
All of these proteins function as a rapid multienzyme complex, therefore, their actions must be thought of as coupled together.
|
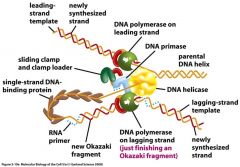
-DNA helicase opens the DNA helix at the front of the replication fork.
-SSB proteins bind the single strand to prevent re-annealing or hairpin loops. -The DNA polymerase of the leading strand operates continuously at the replication fork with the assistance of a sliding clamp. -The DNA polymerase of the lagging strand restarts at short intervals using the discontinuous assistance of a DNA primase to create RNA primers and a clamp loader to pre-load a sliding clamp to each primer. -The replication fork is followed by the enzymes responsible for primer excision and DNA ligase. |
|
|
What is DNA topoisomerase I?
|
-reversibly break phophosdiester bonds
-allows double helix to swivel and relieve tension -Creates and then reseals a single-strand break in the backbone of the double-stranded DNA ahead of the replication fork -Allows both sections of the single strand to rotate freely -uses built up tension and the other strand as a swivel point. |
|
|
What is DNA topoisomerase II?
|
-Creates and then reseals a double-strand break
-This allows a separate DNA molecule crossing over the first to pass through unobstructed. |
|
|
What is a replication origin and what type of bases are these areas usually rich in?
|
Sites at which DNA strands are first separated
Usually AT rich because easier to open. RNA primer synthesis then takes place. |
|
|
What is the function of initiator Proteins?
|
assist in initiating replication by breaking hydrogen bonds to separate strands of DNA.
|
|
|
Are replication origins activated in clusters and how many units apart are they?
|
-activated in clusters
-Origins of Replication are thousands of nucleotides apart. -Replication forks always open in pairs (Replication Bubbles) -DNA Synthesis always occurs during S phase |
|
|
What is the prereplicative complex?
|
Once helicase loading proteins (Cdc6 and Cdt1) are produced, they bind the origin of recognition complex-DNA to form the prereplicative complex
|
|
|
Through what time in the cell cycle of eukaryotic cells is the origin of recognition complex bound?
|
bound through most of the cell cycle
|
|
|
What triggers the passage from G1 to the S phase of cell cycle?
|
Cdks (protein kinases phosphyorylating other proteins) are activated which then dissociate the helicase loading protein.
In turns activates helicase which then unwinds DNA at the origin. DNA polymerases and the remaining replication proteins are then loaded. |
|
|
What is found in the replication fork?
|
the replication machinery
|
|
|
Does eukaryotic DNA or prokaryotic DNA replicate more slowly? And why?
|
eucaryotic
-activated in clusteres called replication units -individual orgines of replication are spcaed tens of thousands of nucleotides apart -replication forks always open in pairs, creating replication bubbles DNA synthesis always occurs during the S phase of the cell cycle and the entire genome is completely replicated -replication units are activated at distinct times throughout S phase and heterochromatin is replicated late in S phase suggesting chromatin condensation is related to timing of replication |
|
|
What bases does the origin recognition complex bind to initiate replication in yeast and prokaryotes?
|
mostly As and Ts
|
|
|
How many histones are required after replication?
|
Equal amounts of new histones as DNA mass are required after replication.
To solve this, humans have about 20 copies of each histone gene from which protein is synthesized in S phase. |
|
|
What type of proteins assist replication forks in moving through parental nucleosomes without displacing them?
|
chromatin-remodeling proteins
|
|
|
Give an example of epigenetic inheritance.
|
refers to heritable traits which occur wihtout changes in a nucleotide sequence, histones are one example
|
|
|
List the roles of chromatin-remodeling proteins
|
-Assist the replication forks in moving through parental nucleosomes.
-facilitates nucleosomes distribution to each daughter strand. -Allows for epigenetic inheritance: Heritable traits which occur without changes in nucleotide sequence. Modification of H3 and H4 (example: Methylation) |
|
|
Describe histone replication.
|
After Replication:
-Equal amounts of histone protein as DNA needed. -Synthesized in S phase from 20 copies of each histone gene. -Histone chaperones bind basic histones and distribute them to fill in holes. |
|
|
Why does telomerase exist.
|
No room to produce RNA primer for last lagging strand, telomeres are segments of DNA at the end of linear strands, composed of many repeating bases.
|
|
|
What does telomerase do?
|
enzyme which recognizes the telomere sequence and elongates it after each cell division, elongates in 5' to 3' direction, utilizes an RNA subunit. Purpose is to protect the overhang from degredation.
|
|
|
What has indefinite polymerase activity?
|
yeast
|
|
|
What happens each time a cell divides to telomerase?
|
Cells lose telomerase through each divison.
Eventually cells become senescence. |
|
|
What is senescence?
|
means that cells can no longer divide due to defects near the ends of chromosoms
|
|
|
What is the Hayflick limit?
|
cells have a limited life span
|
|
|
What are the HeLa cells?
|
cells cultured from Henrietta Lacks who died of cervical cancer, immortal cell line that was used to develop the polio vaccine.
|
|
|
Explain the reasons for cell senescence and immortalization.
|
with every cell division, telomerase is lost, eventually cells become senescence.
immortalization can occur if telomerase production is indefinite. |
|
|
How many nucleotides are lost each time a cell divides?
|
100-200
|

