![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
65 Cards in this Set
- Front
- Back
|
What's the difference btwn A-DANA and B-DNA?
|
A is shorter and wider (dehydrated) and has a C-3' sugar pucker
B is longer and narrower with a C-2' sugar pucker (generally cellular DNA is Bform) |
|
|
What causes the major and minor grooves?
What is the significance of the H-bonding the grooves can form? |
base pairs are not diametrically opposite eachother. Major grove has the bigger side of the base pair (Thy-Methyl) and minor groove has the smaller side (pyrimidine O-2 and purine N-3)
Significance of grooves' side and H bond potential is how proteins can interact with specific DNA sequences. |
|
|
What is a good H bonder in the minor groove?
What is a good H bonder in the major groove? |
N-3 of G or A (minor groove)
N-7 of G and A (major groove) |
|
|
Is DNA perfectly uniform?
|
No way. There are ranges (28-42 degrees vs. the 36 degrees of watson crick model) of degrees of coil and they have local deviations
There can be: propeller twisting Z-DNA Supercoiling |
|
|
What is propeller twisting?
|
base pairs are not perfectly coplaner and the amount of this depends on the base pair sequence.
|
|
|
What is Z DNA?
|
left handed instead of right handed double helix.
occur in short sequences of oligonucleotides that alternate (dCGCGCGCG) Need high salt concentration to alleviate static repulsions, but these guys are present in cells so they must be there to perform a specific function where it doesn't have to be so super salty. |
|
|
Describe the shape of A, B and Z DNA.
|
A=broadest
B=Intermediate Z=Narrowest |
|
|
Describe the screw sense of A, B and Z DNA.
|
A= right handed
B= right handed Z= left handed |
|
|
what are the grooves like of A DNA?
|
major= narrow and deep
minor= broad and shallow |
|
|
What are the grooves of B DNA?
|
major= wide and deep
minor= narrow and deep Type B's are both deep |
|
|
What are Z DNA grooves like?
|
major=flat
minot = narrow and deep |
|
|
What happens if you unwind B-DNA a couple of turns?
|
when it ligates it can have 2 forms
23 turns and an unwound loop or 25 turns and a negative superhelix. |
|
|
What does supercoiling accomplish?
|
much more compact DNA and it moves faster in electrophoresis.
|
|
|
What is the linking number?
|
# of times a strand of DNA winds in he right-handed direction when the helix is constrained to lie in a plane.
|
|
|
What is a topological isomer (topoisomer)?
|
molecules differing only the LK
|
|
|
How can topoisomers be interconverted?
|
only by cutting one or both strands and rejoining them
This is the role of topoisomerases. |
|
|
What are DNA with the same LK but that are geometrically different?
|
have differing Tw and Wr numbers.
|
|
|
what is twist?
What is writhe? |
Tw = helical winding of DNA strands around eachother
Wr= measure of coiling of axis of double helix = supercoiling |
|
|
what is the optimum energy in the expression for the equation that depicts supercoiling?
|
70% of LK expressed in Wr
30% of LK expressed in Tw |
|
|
Does negative supercoiling indicate getting tighter or looser for topoisomerase activity?
|
Means it's getting looser. Most naturally occuring DNA is negatively supercoiled priming it for strand separation for things like transcription and repair.
|
|
|
What do Type I isomerases do?
|
catalyze relaxation of supercoiled DNA
thermodynamically favored so no ATP necessary. alter the LK, cleaves ONE DNA strand, does its thing (replicate, transcribe or recombine) and reseal |
|
|
What do Type II isomerases do?
|
use ATP to create negative supercoils to DNA to make them more compact.
alter the LK, cleaves both DNA strands, does its thing (replicate, transcribe or recombine) and reseal |
|
|
In the cavity of human Type I topoisomerase what is the crucial residue?
Describe how this enzyme works. |
Tyr 723
DNA is bound to the enzyme cavity. Tyr 723 OH is NU that attacks a phosphate group on DNA making a phosphoester bond and cleaving the DNA Now there is a free 5' end |
|
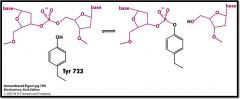
Describe what is happening in this image.
|
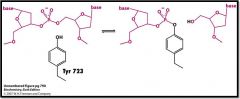
DNA binds to Type I topoisomerase and Try 723 OH acts as a Nu to attack a Pi group on DNA and forming a phosphoester bond and thus cleaving the strand to leave a free 5' end of DNA
|
|
|
What happens once there is a free 5' end of DNA once Type I topoisomerase has cleaved it?
|
Because the freeing of DNA is thermodynamically favorable, the 5' end unwinds some. Then the 5' OH attacks the Try 723 that is bonded to the 3' end of the DNA to reseal the strand.
DNA dissociates from the enzyme. |
|
|
Describe Type II topoisomerase.
|
dimer with 2 internal cavities.
Big cavity has 2 gates (binds G strand tightly and the other gate binds the transported or T strand loosely) both gates have Tyr residues that mimic type I activity. |
|
|
how does one cycle of the activity of Type II topoisomerase effect LK and WR?
|
one cycle decreases the LK by 2
|
|
|
What does novobiocin block?
|
blocks the binding of ATP to bacterial DNA gyrase (the bacterial version of Type II topoisomerase)
|
|
|
What does nalidixic acid and cipro block?
|
block the cleavage and resealing of DNA in bacterial gyrase (bacterial version of type II topoisomerase)
UTI's and other infections (anthrax) |
|
|
What does camptothecin block?
|
human anti-tumor agent
blocks human topo I by stablizing the enzyme when it's Tyr is bound to the DNA and so the strand unwinds way more and endlessly because there is no releasing and resealing. |
|
|
What does a "template-directed enzyme" mean?
|
Each nucleotide coming in first forms a connection with the template base pair and then it is connected to the nucleotide next to it.
|
|
|
Which direction are nucleotides added onto the template?
What is the mechanism? What is needed at the outset? |
added to the 3' end.
the 3' OH of the terminus gets activated by the DNA polymerase to be the Nu that attacks the a-phos group of the nucleoside triphos that is getting added. a primer must be in place with a free 3' OH. |
|
|
What is the Klenow fragment?
|
a part of DNA polymerase I in E. coli
Has 3 domains: 1). "fingers and thumb" wrap around DNA helix 2). "palm" is polymerase activity 3). additional 3'-->5' exonuclease domain (proofreading) |
|
|
How do the 2 metal ions work in DNA polymerase mechanism?
|
One metal ion binds the dNTP and 3' OH of primer (activates the OH to be the Nu)
The other metal ion only interacts with the dNTP The interactions hold and position the stuff properly. Also balance the negative charge that gets created |
|
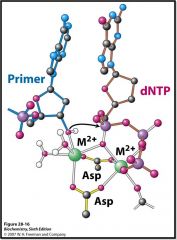
Explain how these metal ions contribute to the mechanism of DNA polymerase.
|
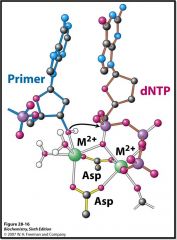
DNA polymerase mechanism:
Two metal ions (Mg2+) One metal ion coordinates 3'-hydroxyl of primer Other metal ion interacts with dNTP Phosphate group of nucleoside triphosphate bridges 2 metal ions Primer hydroxyl attacks phosphate group to form new O-P bond |
|
|
Discuss how H bonding and nucleoside triphos shape contribute to replication specificity.
|
It's not just H bonding or just shape, but a combo of both.
Even if no H bonds can form, an analog to adenosine will still bind thymidine because the shape. |
|
|
Why is shape so important in DNA polymerase activity?
|
minor groove H-bonding of polymerase ensure a properly spaced base pair has formed (H-bond acceptors are in same position for all Watson-Crick base pairs)
Uses this measure to make sure of a proper fit. If the H bonds don't happen the dNTP gets kicked out. |
|
|
What are 3 features of DNA polymerase that ensure fidelity of DNA replication?
|
minor groove H bonding
Shape of dNTP "finger" conformational change that clamps down on dNTP and makes sure only a properly shaped base will fit |
|
|
What is primase?
H |
RNA polymerase
makes the primer with the free 3' OH for the initiation of DNA synthesis |
|
|
How does primase work?
|
Primase synthesizes a short (5 nucleotide) RNA strand that is complementary to one of the DNA strands
|
|
|
What is the replication fork?
|
site of DNA synthesis
leading strand is synthesized continually in the 5'-->3' direction by DNA polymerases The other lagging strand has to make Okazaki fragments and add them on to RNA primers made by primase |
|
|
What are Okazaki fragments?
How do they get linked? |
small fragments of synthesized DNA that get started on the lagging strand by RNA primer and allow for the 5'-->3' synthesis on this strand.
This means the orignal strand running 3'-->5' is growing too, but the growth is in a 5'-->3' way. DNA ligase links the Okazaki fragments. |
|
|
What is DNA ligase?
|
joins the ends of adjacent Okazaki fragments
creates a phosphodiester bond between 3' OH and one DNA end with the 5' Pi group of another. This bad boy only works on ssDNA at the end of a dsDNA so it cannot just link up 2 ssDNA or make a circle or anything like that EXCEPT for T4 bacteriophage ligase which can put 2 blunt ends together |
|
|
What are helicases?
|
use ATP hydrolysis to separate DNA strands
|
|
|
What is bacterial helicase PcrA?
|
extensively studied helicase which gives us an idea how they work.
4 domains A1--P loop NTPase fold (ATP binding and hydrolysis) (bind ssDNA) A2-- B1--like A1 but no P-loop (bind ss DNA) (does NOT bind nucleotides) B2-- When no ATP is around both A1 and B1 are bound to ssDNA |
|
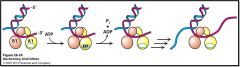
explain the domains and how this helicase PcrA is working to unwind the DNA
|
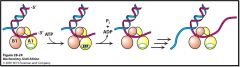
Helicase mechanism
PcrA domains A1 and B1 bind ssDNA ATP binding causes cleft closure between domains Domain A1 slides along ssDNA ATP hydrolysis causes cleft opening DNA is pulled from domain B1 to A1 Process repeated to unwind dsDNA |
|
|
Which direction does the PcrA enzyme work on the ssDNA it binds?
|
3' -->5' direction
|
|
|
What are some characteristics of polymerases with regard to speed and efficiency?
|
high catalytic potency, fidelity and processivity
(processivity is the ability of an enzyme to catalyze many consecutive reactions without releasing its substrate) |
|
|
What is E.coli DNA polymerase III?
|
replicative polymerase
B2 subunit is what gives it that special something. Keeps the polymerase associtated with the DNA DOUBLE helix Looks like a star and acts like a sliding DNA clamp |
|
|
What are clamp loaders?
|
other enzymes that use ATP to pull the DNA polymerase III clamps off the DNA double strand for a sec so the DNA can slide through quickly as its being replicated.
|
|
|
What is SSB?
|
single-strand binding protein. Keeps the unwinding DNA strand (ripped asunder by helicases) from reattaching to them selves
|
|
|
Describe DNA polymerase holoenzyme and how it keeps the 2 sides coordinated while the leading and lagging sides are synthesized.
|
The enzyme has 2 copies of the polymerase core (one works on leading and one works on lagging strands). These are both linked to a central structure which includes the clamp loader complex and the helicase (DnaB)
|
|
|
What is the trombone model?
|
lagging strand loops out in same direction as leading strand. DNA polymerase III working on that strand adds ~1,000 nucleotides and then releases it.
|
|
|
What does DNA polymerase I do?
|
5'-->3' exonuclease activity that removes the primer for ligase to eventually come in and link the fragments.
|
|
|
What is oriC locus?
|
The origin of replication spot for E. coli.
Has 3 identical 13-nucleotide sequences and then 5 AT binding sites that bond with DnaA |
|
|
What is DnaA origin recognition protein?
|
binds to the 5 AT spots on oriC to start this whole party
c-terminus of each of its monomers binds with the 5 spots |
|
|
What is the pre-priming complex?
|
Once a DNA strand gets bound around DnaA other proteins like DnaB helicase and SSB proteins come in to unzip and keep apart the strands. This whole thing is the pre-priming complex.
|
|
|
What is DnaG?
|
the guy that comes to the pre-priming complex and constructs the primer.
|
|
|
What's the difference between prokaryotic and eukaryotic replication origin?
|
pros--significantly smaller genetic info that is circular, has only one origin site.
euk--huge genetic info that is linear, lots of chromosomes, multiple origin sites. |
|
|
What is a replicon?
|
starting site for a replication unit (one of the 30,000 in humans)
|
|
|
If prokaryotes have the specific sequence oriC what ddo eukaryotes have?
|
ORC's
Not specific sequences but rather broadly defined AT sequences where these complexes are assembled ORC's have 6 different proteins homologous to DnaA |
|
|
What are licensing factors?
|
recruiters to the ORC of other proteins like
--Mcm2-7 helicase which is the unzipper --replication protein a (SSB) |
|
|
What is the main processive DNA polymerase in Eukaryotes and how does it get started?
|
DNA polymerase d.
first polymerase a comes in and starts replicating but gets switched out with DNA polymerase d. |
|
|
What are the 3 proteins in DNA repair that are in the MMR systems?
|
MutS (detects mismatch)
MutL (recruits endonuclease) MutH (said endonuclease) |
|
|
What is hereditary nonpolyposis colorectal cancer caused by?
|
defective DNA mismatch repair
There are mutations in the MMR system proteins 70% risk and constitutes 2-5% of all colorectal cancers. |

