![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
40 Cards in this Set
- Front
- Back
|
factors influence enzymatic activity |
1- availability of S and cofactors 2- as P accumulates, rate decreases 3- regulation of E synthesis and decay determine amount of E present 4- activity regulated allosterically 5- regulated covalent modification 6- zymogens, isozymes and modulator proteins play a role |
|
|
allosteric |
other site not active site
|
|
|
covalent modification |
kinase- OH to PO4 phosphatase- take off |
|
|
zymogens |
inactive precursors of enzymes need proteolytic cleavage Pro-insulin- cut off connecting peptide chymotrypsinogen(inactive)- cleave arg15 by trypsin pi chymotrypsin (active)- self digest at Leu13, Tyr 146, Asn 148 alpha- chymotrypsin- active- 3 pieces with disulfide bridges |
|
|
Steps to blood clotting |
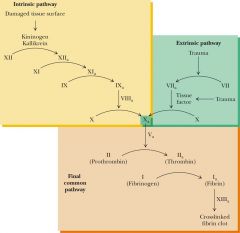
4 carbonyl groups surround Ca2+ to clot of gamma- carboxyglutamate |
|
|
isozyme |
enzymes with 'similar' subunits ex: lactate dehydrogenase- 2 monomer forms A,B- combine differently to make tetramer! A4= liver A3B= muscle A2B2= wbc Ab3= brain, rbc B4= kidney, heart |
|
|
allosteric regulation |
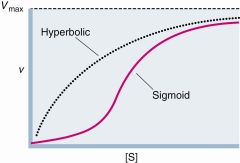
effectors are produced somewhere else feed-forward activators of feedback inhibitors kinetics= SIGMOID activity lower than M-M more extreme off and on |
|
|
Monod, Wyman, Changeux |
allosteric proteins- 2 state- Relaxed, Taut all subunits must be in same state T when no S, S binds tighter to R inhibitor- increase # of T activator- increase # of R- closer to hyperbolic COOPERATIVITY- S binding increase R#, increase sites available ligands (s) are positive homotrophic effectors
|
|
|
heterotrophic effectors |
molecules that influence the binding of something other than themselves |
|
|
Koshland, Nemethy, and Filmer |
ligand triggers conformational change in protein oligomeric- conform in one unit lead to conform in all units NEGATIVE COOPERATIVITY- conform changes cause subunits to adopt conformations with little affinity for ligand SEQUENTIAL MODEL |
|
|
sequential model for allosteric regulation |
positive cooperativity- one unit change then rest will and increase activity no coop- hyperbolic negative coop- one changes then others change to not want the S- decrease in activity |
|
|
differences in KNF and MWC |
MWC- different conformation have different affinities for ligand, ignore ligand-induced conformational changes KNF- based on ligand induced conform change |
|
|
regulate by reversible phosphorylation |
kinase- add phosphoryl- do Ser, Thr, Tyr conserve 260 aa regulated by intrasteric control- regulatory subunit has PSEUDOSUBSTRATE SEQUENCE- mimic the target- block site!! phosphatase- remove phosphoryl |
|
|
cyclic AMP-dependent protein kinase |
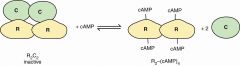
two R bind cAMP cAMP binding releases R from C C are active as monomers release regulatory from catalytic |
|
|
more than phosphorylation |
adenylylation- transfer AMP from ATP to TYr-OH Uridylylation- transfer UMP from UTP to Tyr-OH ADP ribosylation- transfer ADP ribose from NAD+ to Arg redox- reduce Cys-S-S-Cys to Cys-Sh acetylation- transfer acetyl group from acetyl-CoA to Lys E-amino group |
|
|
acetylation |
prominent of e-NH3+ of Lys- change positive amino group to neutral amide add- KAT off- KDACs |
|
|
bnoth allosteric and covalent modification |
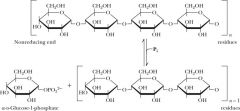
Glycogen phosphorylase- GP GP cleaves glucose from nonreducing ends of glycogen glycogen- G-1-P phosphorolysis rxn PLP is covalently linked allosteric and subunit interface |
|
|
phosphoglucomutase |
G1P-> G6P |
|
|
GP |
dimer of identical 842 subunits each unit has an active site- at center and allosteric effector- near interface phosphorylation site- Ser14 inhibitors- ATP and G6P- theres enough! activator- AMP- need more goes with MWC model conformational change at interface- linked to structural change at active site |
|
|
regulation of GP model |
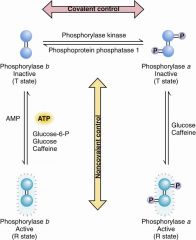
cost 4 ATP per active A-P |
|
|
Krebs and Fischer |
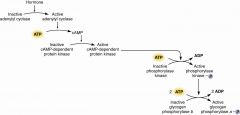
converting enzyme phosphorylase B to A BY covalent phosphorylation |
|
|
1st of krebs |
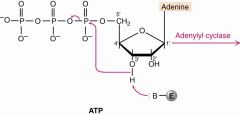
adenylyl cyclase rxn 1- Base attack H of 3'OH, that O go to 1-P kick out PPi (drives rxn) |
|
|
cAMP |
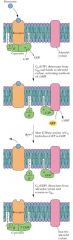
second messenger intracellular agent of hormones hormone stimulates GTP-binding and release Ga, binding of Ga stimulate adenylyl cyclase |
|
|
Al Gilman |
discovered receptor and cyclase saw ATP had trace of GTP |
|
|
Hemoglobin and myoglobin |
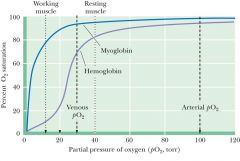
Hb- start 95 to 70- transfer 02 Mb- 98 to 94- more o2 bound
|
|
|
Hemoglobin does.. |
tetramer- 4 hemes carry o2 fe2+ interact with 6 ligands 4- N, 1- imidazole of His F8(sixth helix and 8th residue), 1- o2 |
|
|
Mb structure |
monomeric heme protein cradles the heme Fe2+-ferrous to bind to oxygen if Fe3+- ferric= metmyoglobin- no to o2 |
|
|
o2 alters Mb conformation |
without, Fe is 0.055 nm above the plane with, 0.026 nm- pulled down- little consequence oxygen-storage protein- GREATER AFFINITY torr- hyperbolic
|
|
|
o2 alter hb conformation |
without- 0.06 same as Mb but caused series of conformational changes that goes through subunits must bindd to o2 in lungs(100 torr) and release in capillaries (40 torr)-- cause sigmoid |
|
|
conformational change |
with o2- pull fe into plane along with His f8 ligand- total for Mb= 0.029 nm when deoxy Hb crystalsexposed to o2= shatter Hb- one alpha-beta pair moves by 15 degrees- 0.039 nm!!- rupture salt bridge |
|
|
physiological significance of Hb:O2 |
sigmoid makes it possible binding of o2 affected by h+, Co2, Cl- deoxy Hb higher affinity for H+ than oxy-Hb ph decrease= dissociation of O2 increase HbO2 + H+ --> HbH+ + CO2
|
|
|
Bohr effect |
deffect of H+ on O2 binding of protons diminshes oxygen binding binding of oxygen diminshes protons binding increase pH= best o2 binding |
|
|
Co2 promotes dissociation of O2 |
co2 decrease o2 binding CO2 + h2o--> H+ + HCO3- protons are taken up by Hb |
|
|
tissue-capillary interface |
Co2 hydration- make H+ promote dissociation
|
|
|
lung artery |
bicarbonate dehydration consume H+ so CO2 release and O2 binding |
|
|
2,3 bisphosphoglycerate |
allosteric effector of Hb no 2,3 BPG- oxygen binding- rectangular hyperbola presence- sigmoid 2,3 BPG binds are distant site from Fe to O2 the 5 negative charges interact with 8 positive of 2 Lys, 4 His, 3 Ntermini binds inside central cavity |
|
|
fetal Hb |
lower affiniyt for 2,3 BPG- higher affinity for oxygen differ from adult with gamma chains in place of beta gamma have Ser instead of His at 143- lack two positive charges so BPG bind less tightly |
|
|
sickle cell anemia |
crescent shape RBC pass less freely through capillaries single aa sub in beta chain of Hb Glu at 6 replaced by Val causes aggregation into chainlike polymeric |
|
|
Hb and Nitric oxide |
nitric oxide- neurotransmitter second messenger in signal transduction high affinity for Hb- 10000 tighter than o2 reacts with Sh of Cys 93 of beta to make s-nitroso derivative= Ch2-s-n=o |
|
|
changes in Heme iron upon o2 bindings |
deoxyHb- 6 d electrons of fe2+ has 4 unpaired electrons and one pair four ligands to ring system 1 to histidine F8 iron is paramagnetic- high-spin state bind to o2- three electron pairs, low-spin state change in spin state- allow bond btw Fe and histidine to be perpendicular and shorten 4 N ligands strengthen |

