![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
112 Cards in this Set
- Front
- Back
- 3rd side (hint)
|
Gregor Mendel |
Priest discovers inheritence with pea plants |
|
|
|
Miescher |
Discovered DNA |
|
|
|
Chargroff 48 |
G=c a=t Figured out they had one to one ratio |
|
|
|
Rosalind Franklin and Maurice Wilkins (1950) |
Show DNA is double stranded |
|
|
|
Watson Crick and Wilkins (1953) |
Determine dna's structure |
|
|
|
Fred Griffith (1928) |
Discovers transformation -Live "R" strains (which are normally not lethal) could turn heat killed "S" strains (normally not lethal) into lethal |
|
|
|
Avery, MacLeod and McCarty (1944) |
Figure out DNA is "Transforming Particle" from Griffith's experiment |
|
|
|
Hershey and Chase (1952) |
Figured out DNA is genetic material in T2 bacteriophage by tracking proteins and DNA with radioisotopes. |
|
|
|
What is Central Dogma? |
DNA->Transcription->Translation |
|
|
|
Nucleic acid structure |
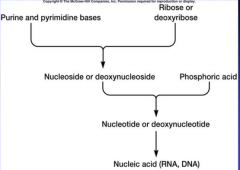
|
|
|
|
Purine or Pyrimidine plus a ribose or dexoyribose turns into what? |
Nucleoside or deoxy nucleoside |
|
|
|
A nucleoside or deoxsy nucleoside plus a phosphoric acids turns into what? |
nucleotide or deoxynucleotide |
|
|
|
Which DNA groove do most mods occur in |
Major |
|
|
|
What is the backbone of DNA? |
Sugar and phosphate |
|
|
|
Prokaryotic DNA characteristics |
circular supercoiled (held together by histone-like proteins) and not bound by a membrane |
|
|
|
Semiconservative replication |
Each copy of DNA contains an old and a new strand |
|
|
|
Replication bubble |
the area of DNA that is being replicated |
|
|
|
Replication Fork |
The area where replication is actually taking place |
|
|
|
Two patterns of DNA synthesis |
1. bidirectional 2. Rolling Circle |
|
|
|
DnaA |
Protein which binds to DnaA box in OriC region and opens up the double strands. |
|
|
|
Organization of eukaryotic DNA |

Linear membrane bound business nucleosomrs |
|
|
|
Bidirectional |
Two forks move in the opposite direction |
|
|
|
Theta structure |
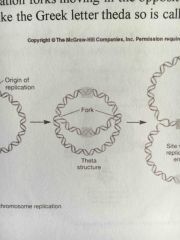
Forms during prokaryotic replication of circular DNA |
|
|
|
How does tooling replication work? |
1. Nick 2. 3' end has growing stand which displaces other stand 3. DNA synthesis occurs in displaced strand |
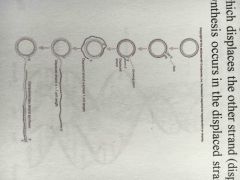
|
|
|
Mechanism of chain growth |
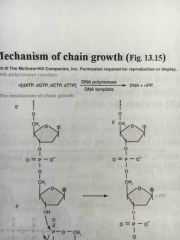
Back (Definition) |
|
|
|
Mechanism of chain growth |
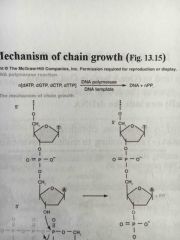
|
|
|
|
Purines |
A and g |
|
|
|
Pyrimidine |
T and C |
|
|
|
DNA structure: |
Doublestranded Complementary Antiparallel |
|
|
|
Two hydrogen bonds |
A and T |
` |
|
|
Three hydrogen bonds |
G and C |
|
|
|
Eukaryoitic Dna organization |
1. Linear 2. Membrane bound 3. Histones (nucleosomes) |
|
|
|
What separates the strands during DNA replication |
Helicase (DnaB) |
|
|
|
DnaA |
A protein which binds to the DnaA box in the OriC region |
|
|
|
DNA gyrase |
Minimizes over-twisting of DNA |
|
|
|
What inhibits DNA gyrase? |
1. Novobiocin 2. ciprofloxacin |
|
|
|
DnaB |
Pre-Primer for Primase |
|
|
|
Primase |
DNA dependent RNA polymerase which attaches to DNA and synthesizes showrt complementary RNAs (about 10 nucleotides) |
|
|
|
SSB |
Single stranded binding proteins attach to each strand to keep them from reannealing |
SSB Strands stay by themselves |
|
|
DNA polymerase III |
Synthesizes leading strand (10 proteins-core synthesizes) (E subuint proofreads) |
Threeding strand |
|
|
What are the limitations of DNA poly III? |
1. Adds nucleotides to pre-existing 3'OH ends ONLY 2. Needs pre-existing 3'OH to begin synthesis 3. needs a template strand |
|
|
|
Lagging strand |
1. Ozaki fragments 2. PrePriming and synthesis of RNA primers 3. Addition of complimentary DNA poly III |
|
|
|
DNA poly I |
Removes teh RNA primers 5-3' exonuclease and replaces it with complementary DNA |
|
|
|
What can proofread during REPLCIATION |
DNA poly 3 |
|
|
|
What is Exonuclease? |
E subunit of DNA Poly III |
|
|
|
What adds methyls to new bases during replication |
Methylases |
|
|
|
What seals the nick |
DNA ligase |
|
|
|
When does replication stop? |
When a ter site is reached |
|
|
|
How does replication stop? |
Tus proteins bind causing replication complex to fall off. |
|
|
|
What part of tRNA does the amino acid attach to? |
3' end attached to Oxygen |
|
|
|
What directions are codons read? |
5' to 3' |
|
|
|
The basic structure of a gene on DNA |
Promoter, leader, coding, trailer and terminator. |
|
|
|
What part of mRNA does the ribosome bind to? |
shine-dalgarno |
|
|
|
Sense strand |
contains coded info |
|
|
|
antisense strand (template) |
Complimentary to sense strand. the beginning of the gene starts at the 3' end and mRNA is synthesized from 5' to 3' end. This sstrand is transcribed |
|
|
|
Recognition site on DNA |
-35 |
|
|
|
RNA polymerase binding site? |
-10 |
|
|
|
Promoter? |
DIRECTS the binding of RNA polymerase |
|
|
|
Leader sequence? |
codes for a segment of mRNA (leader) |
|
|
|
what does the leader code for? |
The shine-dalgarno |
|
|
|
What part of the ribosome binds to shine dalgaron |
16s |
|
|
|
Coding region starts with what? |
3'TAC5' which becomes AUG in the mRNA and codes for the initiation of translation |
|
|
|
Terminator? |
The sequence located after a non-coding sequence (trailer) and codes for termination of transcription |
|
|
|
What is coded for by a large transcript? |
rRNA
|
|
|
|
How is rRNA processed by prokaryotes? |
a ribonuclease cuts up the large transcript into 1-3 tRNAs and a 16s, 23s and 5s |
|
|
|
Conditional mutation |
mutations expressed only under cetain environmental conditions |
|
|
|
Biochemical mutations |
change of the biochemistry of the cell |
|
|
|
Prototroph |
bacterium that can grow on minimal medium |
its a pro, and can synthesized its own **** |
|
|
Auxotroph |
bacterium which requires a nutrient supplement to grow (amino acids) |
|
|
|
Mutations are either blank or blank? |
Spantaneous (natural) or induced (mutagens) |
|
|
|
Transition mutation |
purine to purine
pyrimidine to pyrimidine |
|
|
|
Transversion mutation |
purine to pyrimidine |
|
|
|
intercalating agents |
distort DNA lead to indels (ethidium bromide) |
|
|
|
true reversion |
An exact reverse mutation |
|
|
|
suppressor mutation |
a mutation which generates wild type |
|
|
|
intragenic suppression |
within the same gene |
|
|
|
extragenic suppression |
somewhere else |
|
|
|
nonsense suppression |
tRNA mutation |
|
|
|
physiological suppression |
defect in one pathway is overcome by a mutation in a another pathway |
|
|
|
Silent mutation |
no visible effect |
|
|
|
missense mutation |
change in amino acid occurs |
|
|
|
nonsense mutation |
mutation makes a nonsense codon (STOP) |
|
|
|
Whats a test to test mutations |
Ames test |
|
|
|
Mismatch repair |
1. post replication 2. DNA poly three can do it with its mismatch repair 3. Dnay methylation (methyl directed repairs) 4. Mut proteins |
|
|
|
Excision repair |
1. nucleotide excision repair (uvrABC endonuclese) 2. base excision repair (glycolysase and AP endonuclease) removes damage and unnatural bases |
|
|
|
Removal of lesions |
Direct repair (photoreactivation-photolyase splits thymine dimers OR alkyltransferases) |
|
|
|
Akyltransferase |
Removal of methyls or alkyls which damaged bases...Directly |
|
|
|
Recombination repair |
both bases of a pair are missing, gap exist RecA protein mediated |
|
|
|
What happens to protein levels when damage occurs? |
They are increased in expression because bound RecA destroys lexA which is a repressor |
|
|
|
core enzyme of RNA polymerase |
1. 2 alpha subunits - recognizes promoter, assembles core enzyme 2. a beta subunit - binds ribonucleotide substrates 3. R' subunit - binds to RNA |
|
|
|
Sigma subunit of RNA polymerase |
helps RNA polymerase holoenzye recognize promoter |
|
|
|
SIGMA + CORE = |
holoenzyme |
|
|
|
Prokaryotic termination |
rho dependent or not |
|
|
|
whats the intrinsic terminator |
Stem and loop (tryptophan thing u-rich areas) |
|
|
|
Whats polycistronic? |
multiple coding regions...bacteria only |
|
|
|
Eukaryotic rna polymerases?
|
RNA polymerase I II and III |
|
|
|
Eukaryotic poly-A tails |
A type of post-transcriptional processing 7-methylguanosine |
|
|
|
Where does translation occur in eukaryotic cells |
cytoplasm |
|
|
|
where does splicing occur in eukaryotic cells |
nucleus |
|
|
|
whats polysome? |
mRNA plus a few ribosomes |
|
|
|
Transcription and Translation are coupled in: |
bacteria |
|
|
|
Structure of tRNA |
1.acceptor stem 2. TyC arm 3. Variable arm 4. Anticodon arm 5. D arm |
|
|
|
Initiaon in prokaryotes |
forming 70s initiation complex |
|
|
|
elongation process in bacteria |
adding amino acids to growing polypeptide |
|
|
|
termination in bacteria |
end of translation |
|
|
|
what is the main enzyme in elongation |
peptidyl transferase - transpeptidation |
|
|
|
Whats RF and when are they used |
Release factors are proteins necessary for termination |
|
|
|
What helps proteins fold? |
Chaperones |
|
|
|
SPlicing in microbes? |
Some microbial proteins are modified by the removal of an internal part of the polypeptide before folding of the protein |
|
|
|
What are the equivalents of exon and introns in bacteria? |
extein and inteins |
|
|
|
protein complex types in membrane of gram negatives outer membrane |
II and V transport proteins out of Sec-dependent or Tat pathway I and III are Sec-independent IV - conjugation |
|
|
|
Which type of membrane protein in G- is for conjugation |
Type 4 |
|

