![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
41 Cards in this Set
- Front
- Back
|
Protein definition: a series of _____ linked by _____ bonds (aka _____ bonds) |
Amino acids Covalent/peptide bonds
Formed by dehydration (bond formed btw COO and NH3; O from COOand H2 from NH3 leaves); There will be a free amino end and free carboxyl end on ends of polypeptide chain
|
|
|
The _________ of a protein determines its form an function |
Linear sequence |
|
|
List the four structures of protein categorization |
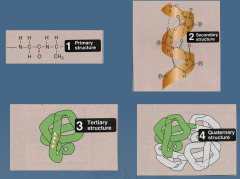
Primary Secondary Tertiary Quaternary |
|
|
Define the primary structure |
Linear sequence of amino acids Can be determined using experimental techniques |
|
|
The peptide bond is a special type of _____ bond that can be broken with _______ |
Covalent
Strong acid or base @ high temperature |
|
|
List the 3 characteristics of a peptide bond |
1. Lack of rotation around the N-C=O bond (acts like a double bond). There is rotation around the C--alpha bonds (carbon next to the carbon in the peptide bond)
2. Trans configuration (due to bulky R groups)
3. Uncharged but polar (peptide bonds neither accept nor accept protons @ pH 2-12; H-bond involved) |
|
|
Formation of a peptide bond involves a ____ reaction |
Dehydration |
|
|
Read peptides from free_____ group on the left to the free ____ group on the right |
Amino
Carboxyl |
|
|
How would you read a peptide composed of: Valine-glycine-leucine |
valyl-glycyl-leucine |
|
|
What are the 3 steps of determining the amino acid composition of a polypeptide? |
1. Acid hydrolysis (strong acid, 110C, 24 H)
2. Cation exchange chromatography (neg AA bind to positive column; wash out with incr ionic/pH solutions)
3. Quantitative analysis (heat separated AA with ninhydrin; spectroscopically measure the resulting purple color) - to get CONCENTRATION of each type of AA for relative composition
|
|
|
Edman's reagent: 1. What is the chemical 2. What does it cleave 3. What is it for 3. Limitations? |
1. Phenylisothiocyanate 2. N-terminus amino acid 3. Sequencing polypeptide from N-terminus 4. Only for polypeptides with 100 or less AA; otherwise inaccurate |
|
|
Name and describe two-step process of sequencing polypeptides via fragmentation (AAs over 100) |
1. Enzymatic - Trypsin: digestive pancreatic anzyme; cleaves carbonyl end of lysine and arginine
2. Chemical - Cyanogen bromide: cleaves at carbonyl end of Methionine (Met) <<Chocolate Baked Mousse>>
LAM - lysine arginine methionine Overlap fragments to sequence |
|
|
How else can you figure out primary structure? |
DNA Sequencing - look at nucleotides -3 nucleotides = a codon; -Each codon will correspond to an amino acid |
|
|
Define multimeric proteins |
Protein with more than one polypeptide |
|
|
How do you separate multimeric proteins into individual polypeptides? |
Denaturing agents |
|
|
Denaturing agents disrupt ______ bonds; 3 examples are _____ |
Non-covalent
1. Urea 2. Guanidine HCl 3. Performic acid (breaks S-S bonds) |
|
|
Define and list structures (3) of secondary structures of proteins |
Regular, 3D arrangements found within a protein as adjacent amino acids interaction.
1. Alpha helices 2. Beta sheets 3. Beta bends |
|
|
a-Helix is a ____ structure with its R groups extending (in/out)wards.
There are ____ amino acids per turn |
Spiral Outwards 3.6 |
|
|
Compare and contrast KERATIN v. HEOMGLOBIN |
Keratin (100% a-helix, rigid bc S-S bonds) Hemoglobin (80% a-helix, globular and flexible) |
|
|
a-Helix hydrogen bonds: 1. What do they connect? 2. Are they weak or strong? |
1. Carbonly of one AA + NH of another AA 4 residues ahead.
2. Weak individually, but strong collectively (most contributing bond type in secondary structure) |
|
|
List nine AA that disrupt a-helix |
BULKY OR CHARGED AAs: Proline (bulky imino group) Charged amino acids (Glutamate, Histidine, Lysine, Arginine, Aspartate) Tryptophan (bulky R group) Valine and Isoleucine (b-Carbon branching)
Partially Gregarious Homer Licked Ancient Archie's Then Vibrant Igloo |
|
|
What is the appearance of b-sheets? |
Pleated |
|
|
Can beta sheets have more than one polypeptides? What about alpha helices? |
Yes
No |
|
|
Hydrogen bonding in b-sheets (2) |
All peptide components engaged in H-bonding
Perpendicular in b-sheets |
|
|
What are the two forms of b-sheets? |
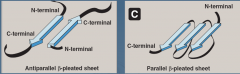
Antiparallel - H bonds form in opposite directions
Parallel - H bonds form in same direction
|
|
|
What is an amyloid protein?
Disease association? |
Fibrous protein composed of b-sheets
Twisted b-sheet amyloid proteins accumulate to cause Alzheimer's disease (identical to silk fibrils) |
|
|
Beta bends:
1. ______ direction of polypeptide chain 2. What is its shape? 3. Consists of _____ AA, usually ___ and ___ 4. What does it usually connect? |
1. Reverses 2. Compact, globular 3. 4, Proline or Glycine 4. Successive strands of antiparallel b-sheets |
|
|
What are non-repetitive structures?
|
Parts of globular protein that isn't a-heilces or b-sheets
Loops and coils
Not random, but not as regular as a-helices or b-sheets |
|
|
Define and list 4 examples of supersecondary structures |
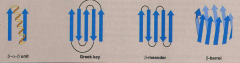
Formed by packing the R groups of adjacent secondary structures.
1. B-a-B unit 2. Greek key 3. B-meander 4. B- barrel
|
|
|
Tertiary structure:
1. _____ + _____ = _____ (functioning unit of 3D structure)
2. 3D structure determined by_____ |
1. Secondary structures + motifs = domains 2. AA sequence |
|
|
Domains: 1. Fundamental ___ and ___ unit 2. Core of domain composed of ___ 3. Each domain is structurally ___ |
1. Functional and structural 2. Supersecondary structures 3. Independent |
|
|
List the 4 stabilizing interactions |
1. Disulfide bonds (most stabilizing interaction in tertiary structure) 2. Hydrophobic interactions 3. Hydrogen bonding 4. Ionic interactions |
|
|
Disulfide bonds: 1. What does it bond 2. What chemical can (reduce/oxidize)s it? |
1. Covalently link the -SH groups on cysteine molecules (cysteine residue) 3. B-mercaptl ethanol REDUCES S-S bonds |
|
|
Hydrophobic interactions: 1. What does it involve? 2. Where is it (outside/inside) protein? |
1. Non-polar R groups 2. Inside if protein in hydrophilic environment; outside if protein in hydrophobic environment. |
|
|
Hydrogen bonding: 1. What does it involve 2. What does it do for the overall protein (other than stabilize)
|
1. -OH or -NH containing R groups + carboxyl/carbonyl groups of other AA
2. Helps solubilize protein in aqueous solutions |
|
|
Ionic interactions: 1. What does it bond |
1. Neg and pos charged R groups to stabilize protein |
|
|
Chaperons: 1. What does it do 2. What is it also known as 3. How does it do what it does? |
1. Aid folding of proteins during translation 2. Pcb proteins - polypeptide chain binding proteins 3. Reads signals on polypeptides itself for how to fold it |
|
|
Quaternary structure: 1. Polypeptides subunits are arranged and held together by ______ interactions 2. Do subunits work together or independently? 3. Nomenclature? |
1. Non-covalent 2. Either or 3. Dimeric, trimeric, or multimeric. |
|
|
Define denaturation & 6 denaturing agents.
Do proteins fold back to native state after denaturation? |
Unfolding and disorganizing of a protein's structure.
1. Urea 2. Heat 3. Mechanical mixing 4. Strong acids + bases 5. Detergents 6. Heavy metal ions (Pb, Hg)
Rarely |
|
|
Prion molecules contain _____ instead of _____ and cause normal proteins to do the same |
beta sheets instead of alpha helices
Gave you a "B" instead of an "A"
*Happens exponentially because infected protein dissociates in two and infects 2 more proteins |
|
|
What (4) diseases does the prion protein cause? |
1. TSEs (transmissible spongiform encephalopathies) 2. Creutzfeldt-Jakob disease 3. Scrapie (sheep) 4. Mad cow disease (bovine spongiform encephalopathy) |

