![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
12 Cards in this Set
- Front
- Back
|
1. What were Meselson and Stahl studying? |
DNA replication. |
|
|
2. How many origins of replication are there in humans? Bacteria? |
Many are found in humans and usually only one is found in bacteria. |
|
|
3. What is the difference between homologous recombination and NHEJ? |
Non-homologous end joining repairs doublestrand breaks, however, it is error prone. May cause a loss or gain insequence. Not always precise Homologous recombination is flawless/seamless repair of a double strand break. |
|
|
What’s a retrovirus? |
A virus that is composed of RNA. |
|
|
4. True or false homologous recombination is very precise in repair double strand breaks |
True |
|
|
5. What are mobile genetic elements? Are any of these genetic elements in the human genome? |
Genomes have mobile genetic elements called transposons. Transposones encode enzyme that allow for movement ofthe DNA sequence (transposase) and encode for antibiotic resistance |
|
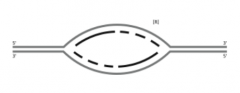
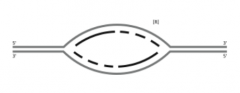
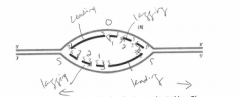
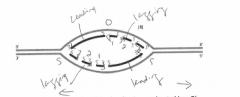
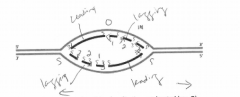
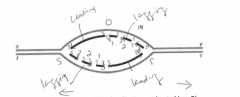
A. On the figure, indicate where the origin of replication was located (use O). |
|
|
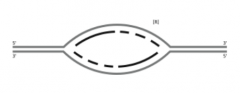
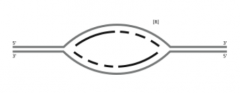
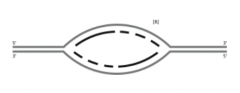
![B. Label the leading-strand template and the lagging-strand template of the right-handfork [R] as X and Y, respectively.](https://images.cram.com/images/upload-flashcards/53/43/73/23534373_m.png)
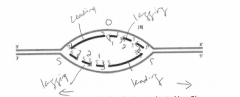
B. Label the leading-strand template and the lagging-strand template of the right-handfork [R] as X and Y, respectively. |
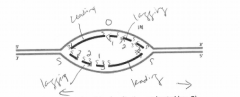
|
|
C. Indicate by arrows the direction in which the newly made DNA strands (indicated bydark lines) were synthesized. |
5' -> 3' |
|
D. Number the Okazaki fragments on each strand 1, 2, and 3 in the order in which theywere synthesized |
|
|
E. Indicate where the most recent DNA synthesis has occurred (use S). |
|
|
F. Indicate the direction of movement of the replication forks with arrows. |
|

