![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
115 Cards in this Set
- Front
- Back
|
ligand
|
binds to receptors
|
|
|
endocrine
|
communicates with distant target and releases land into the blood
|
|
|
paracrine
|
communicates with distant target through tissue coordination and doesn't release ligand into the blood
|
|
|
cell to cell contact
|
cells permanently attached with no diffusable ligand.
|
|
|
autocrine
|
produces a ligand that directly feeds back on the cell that produced it
|
|
|
cell surface receptor
|
when the ligand binds, it causes the protein to change shape and results in the cytoplasmic domain aquiring physiological action
|
|
|
intracellular receptors
|
binds to hydrophobic ligand that can cross the membrane
|
|
|
intracellular receptors
|
-bind to hydrophobic ligands
-5-10k/cell -800 amino acids long -3 separate domains |
|
|
mechanism of intracellular receptors
|
1) ligand binds and changes protein shape
2) imports to nucleus 3) binds to specific DNA sequence 4) N-terminus promotes transcription of that gene 5) mRNA's exported 6) translation |
|
|
cell surface plasma membrane receptor types
|
ion channel linked (ligand-gated channels)
G-protein linked enzyme linked |
|
|
curare
|
plant toxin that blocks nicotinic cholinergic receptors (ligand-gated Na+) and results in paralysis
|
|
|
what can G proteins bind to?
|
gtp or gdp
|
|
|
signal transduction steps
|
1)ligand binds to receptor and it changes shape
2) receptor alters interaction with protein 3) G-protein releases GDP and bonds to GTP instead which activates the G-protein |
|
|
alpha subunit
|
binds guanine nucleotides
|
|
|
beta gamma subunit
|
interacts with receptor
|
|
|
active G-protein function
|
-turn on enzyme
-open/close ion channels -promote production of intracellular signal molecules |
|
|
things g- protein is linked to
|
An isoprene and fatty acid
|
|
|
active g-protein function
|
-turn on an enzyme
-open/close ion channels -promote production of intracellular signal molecules |
|
|
reasons to have multiple steps in signal cascades
|
1) amplify the signal
2) precise regulation |
|
|
what does g-protein do after inactivation?
|
alpha-gdp binds to beta gamma subunits to form heterotrimer complex
|
|
|
Gs
|
- increases adenyl cyclase, which increases cAMP
- opens Ca2+ channel, which increases Ca2+ -closes Na+ channel, which creates voltage |
|
|
Gi
|
-decreases adenyl cyclase, which results in decreased cAMP
- increases K+ channel, which creates a voltage |
|
|
Gq
|
increases phospholigase C, such brings IP3 and DAG
|
|
|
Gt
|
increases phosphodiesterase, which results in decreased cGMP
|
|
|
PKA synthesis
|
1) receptor activates Gs
2) alpha-GTP detaches and activates adenyl cyclase 3) adenyl cyclase converts ATP to cAMP 4) cAMP binds to kinase |
|
|
PKA subunits
|
2 are catalytic and 2 are regulatory
|
|
|
role of PKA
|
phosphorylation
|
|
|
types of pka
|
-serine/threonine kinase
-tyrosine kinase (doesn't phosphorylate it though) - Sugar kinase -lipid kinase |
|
|
adipose
|
ligand: epinefrin
results in breakdown of triglyceride |
|
|
liver
|
ligand: glucagon
results in glycogen breakdown |
|
|
ovarian follicle
|
follicle stimulating hormone that results in estrogen synthesis
|
|
|
Bone cells
|
ligand: parathyroid hormone
results in Ca2+ absorption |
|
|
intestine epithelio
|
-PKA phosphorylates Cl- channel
-NaCl flux into intestinal lumen |
|
|
cholera toxin
|
enzyme that modifies alpha subunit of Gs G-protein, which results in loss of GTPase activity. It's constantly bound to GTP, which increases AC, cAMP, PKA, and the Cl- channel's always open
|
|
|
liver
|
glycogen storage, short-term energy storage, glucagon, increased pka, and pka phosphorylation
|
|
|
glycogen phosphorylase kinase
|
phosphorylates and activates glycogen phosphorylase
|
|
|
cAMP response element (CRE)
|
DNA sequence
|
|
|
CRE binding protein (CREB)
|
will only bind to CRE when CREB is phosphorylated
|
|
|
bordetella pertussis
|
-enzyme that's a toxin
-modifies the Gi -can't release GDP -Gi is permanently inactivated -results in more cAMP |
|
|
Calcium regulation
|
1) 7-transmembrane receptor
2) activates Gq 3) alpha-GTP 4) Gq activates phospholipase C 5) PLC cleaves Phosphatidyl inositol 4,5 biphosphate, which increases IP3 in the cytoplasm 6) IP3 can bind to IP3 receptors, which is a ligand-gated Ca2+ channel on the ER 7) increases Ca2+ in the cytoplasm |
|
|
conventional protein kinase C
|
requires Ca2+ DAG and phosphatidyl serine, which drives it to the plasma membrane
|
|
|
novel protein kinase C
|
doesn't require Ca2+, but it needs DAG
|
|
|
atypical Protein kinase C
|
doesn't require Ca2+ or DAG
|
|
|
CaM (calcium/calmodulin complex) kinase
|
autophosphorylates (maintains activity even if there's not a lot of Ca2+
|
|
|
calmodulate
|
-kinase
-acticates contraction in smooth muscle -acticates synaptic proteins -activates transcription factors |
|
|
calcium
|
1) receptor
2) G-protein 3) plc 4) phosphatidyl inositol + 2 P --> IP3 + DAG 5) IP3--> opens Ca2+ channels on ER 6) Ca2+ flux from ER--> cyto...acticates PKC and CaM kinase |
|
|
cytoplasm Ca2+ concentrations
|
10^-7 M
|
|
|
ER Ca2+ concentrations
|
10^-3 M
|
|
|
ECF Ca2+ concentration
|
10^-3 M
|
|
|
Ras superfamily
|
-not linked to 7-transmembrane domain receptors
- small - over 100 members - regulatory mechanisms consist of GTPase activating protein, which turns off G-protein |
|
|
GAP (GTPase activating protein)
|
turns G-protein off
|
|
|
GEF (guanine nucleotide exchange factor)
|
turns G-protein on
|
|
|
5 branches of the family
|
1) Ras- transcriptional activity (related to cancer)
2) Rho- cytoskeletal organization 3) Rab- vesiculation membrane fusion 4) Ran- import/export from nucleus 5) Arf- vesiculation fusion of membranes |
|
|
receptor tyrosine kinase
|
- common ligands are growth factors
- ligand binds and receptor forms a dimer |
|
|
SH2 binds to...
|
phosphate
|
|
|
cycle
|
sos (guanine nucleotide exchange factor)--> Ras (GTP)--> Raf (kinase)--> MEK (phosphate)--> Erk (phosphate)--> transcription factors change erk to TF-PO4--> imported to nucleus --> transcription--> new mRNA--> new proteins
|
|
|
AKT
|
enzyme that results in tissue development and is dependent on cell environment
|
|
|
phospholipase D
|
R group phosphatidic acid
|
|
|
phospholipase C
|
cuts between phosphate and fatty acids to get IP3 diacylglycerol
|
|
|
PLA2
|
releases an unsaturated FA (eicosanoid), which is used to produce paracrine molecules
|
|
|
prostaglandins
|
associated with inflammation and activated by cycloxygenase
|
|
|
PLA1
|
cuts off saturated fatty acid
|
|
|
cyclo-oxygenase
|
comes from eicosanoids. leads to prostaglandins (smooth muscle control inflammation)
|
|
|
lipo oxygenase
|
from eicosanoids. leads to leukotrienes (smooth muscle control)
|
|
|
aspirin
|
non -steroidal anti-inflammatory
|
|
|
corrisol
|
promotes transcription of a protein that blocks PLA2
|
|
|
ATP energy release
|
7.3 kcal/mol
|
|
|
cellular respiration steps
|
1) substrate --> oxygen
--> organic fuel 2) fuel is oxidized, which releases energy and powers ATP production 3) products are ATP, CO2, and H2O |
|
|
fuel
|
sugars, fatty acids, and amino acids
|
|
|
glycolysis
|
- oxidation
-lytic event - small amount of ATP |
|
|
kreb Chile
|
- utilizes glycolytic products
- mitochondrial matrix - oxidation - small amount of ATP |
|
|
two ways to make ATP
|
1) substrate level phosphorylation (phosphotylator needs more energy than ATP)
2) oxidative phosphorylation |
|
|
phosphenol pyruvate delta G value
|
-14.8 kcal/mol
|
|
|
1, 3 diphosphoglycerate delta G value
|
-11.8 kcal/mol
|
|
|
phospjocreatine (muscle) delta G value
|
-10.3 kcal/mol
|
|
|
glucose-6-phosphate delta G value
|
-3.3 kcal/mol
|
|
|
regulation of glycolysis
|
1) regulated steps tend to be those with a high -delta S value
2) specific control: a) glucose concentration (3-4 mM) b) hexokinase c) phosphofructo kinase |
|
|
pyruvate reduction
|
leads to lactic acid in us and ethanol in yeast
|
|
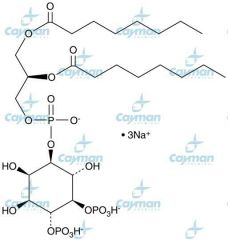
|
phosphatidyl inositol 4,5 biphosphate
|
|
|
creb cycle
|
CREB--> CREB-PO4--> goes to nucleus--> binds to CRE--> transcription
|
|
|
F0F1-ATPase
|
makes ATP
|
|
|
pyruvate after oxidation and loss of CO2
|
acetyl-CoA
|
|
|
pyruvate dehydrogenase
|
-enzyme that catalyzes oxidation of pyruvate
- regulated allosterically - inhibited by NADH and ATP - activated by high NAD+ |
|
|
triglycerides
|
adipose tissue that liberates FAs
|
|
|
fatty acids
|
supplies acetyl-CoA by beta- oxidation
|
|
|
kreb cycle
|
feeds 2C acetyl subunits into the cycle and the cycle will release 2 CO2 electrons
|
|
|
2 acetyl-CoA
|
turns off the kreb cycle
|
|
|
kreb cycle outcome
|
4 CO2
2 NADH 2 FADH2 2 ATP |
|
|
glucose energy harvested
|
680 kcal/mol
|
|
|
4 ATP energy harvested
|
30 kcal
|
|
|
NADH energy harvested
|
52.6 kcal/mol
|
|
|
FADH2 energy harvested
|
43.4 kcal/mol
|
|
|
electron transport system
|
1) NADH dehydrogenase complex accepts electrons from NADH and uses that energy to pump 4 protons across the inner mitochondrial membrane
2) NADH dehydrogenase donates electrons to ubiquinone (co-enzyme Q and membrane lipid) 3) Co-enzyme Q donates electrons to cytochrome b-C1 complex and uses that energy to pump 4 protons across the inner mitochondrial membrane 4) cytochrome b-C1 complex donates electrons to cytochrome C 5) cytochrome C transfers electrons to cytochrome C-oxidase complex, which uses every to pump 2 protons across the inner mitochondrial membrane 6) cytochrome C oxidase transfers electrons to oxygen |
|
|
NADH
|
10 protons pumped across the inner mitochondrial membrane
|
|
|
FADH2
|
6 protons pumped across the inner mitochondrial membrane and bypasses the NADH dehydrogenase
|
|
|
F0 segment
|
proton channel
|
|
|
F1 segment
|
attached to the matrix side and spins as protons go through matrix
|
|
|
kreb cycle produces...
|
4 CO2
6 NADH 2 FADH2 2 ATP |
|
|
F0F1 ATPase conformational states
|
1) high affinity binding for ADP and phosphate
2) condensation state 3) low affinity ATP binding |
|
|
every 120 degree turn of F0F1 ATPase results in...
|
1 ATP released. turn is promoted by H+
|
|
|
NADH
|
energy to pump 10 H+ across inner mitochondrial membrane and produce 3 ATP
|
|
|
FADH2
|
pumps 6 H+ across the membrane to make 2 ATP
|
|
|
10 NADH
|
produces 30 ATP
|
|
|
2 FADH2
|
makes 4 ATP
|
|
|
glycolysis
|
results in 2 ATP
|
|
|
theoretical vs actual yield of ATP
|
theoretical: 38
real: 27-28 |
|
|
ways to lose energy in ATP synthesis
|
1) ADP goes in and H+ goes out
2) pyruvate and H+ go in the matrix 3) phosphate and H+ go in the matrix 4) creatine and H+ go in the matrix (only happens in muscle) 5) transport of glycolytic NADH electrons into the matrix |
|
|
ways to lose energy
|
1) transport ATP into matrix and H+ out
2) pyruvate and H+ go in at the same time 3) phosphate and H+ go in at the same time 4) creatine and H+ go in at the same time 5) transport of glycolytic NADH electrons into the matrix |
|
|
energy that can't be used to power the F0F1 ATPase
|
ADP in
ATP out phosphate in pyruvate in NADH electrons in |
|
|
Brown fat
|
highly vascularized, lots of mitochondria, and produces a protein called thermogenin is m the mitochondria
|
|
|
artificial protonophore
|
H+ channels
|
|
|
DHP
|
artificial H+ channel that increases oxygen consumption, ETS, fuel consumption, and takes down ATP production
|
|
|
cyanide
|
electron transport blocker. blocks the cytochrome C
|
|
|
result of everything backing up
|
- all become reduced upstream
- no H+ pumping - electrochemical gradient collapses - no oxidative ATP produced - death |
|
|
PI-3 kinase
|
phosphorylates C #3 on inositol
|

