![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
209 Cards in this Set
- Front
- Back
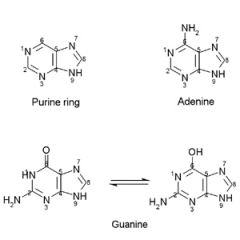
Adenine & guanine have ___ ring stuctures.
They are found in: |
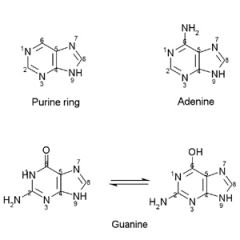
Adenine & guanine have purine ring stuctures.
They are found in: both DNA & RNA |
|
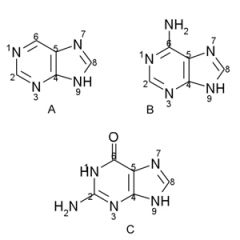
Name the structures and if they are found in DNA and/or RNA.
|
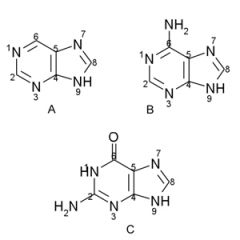
A. Purine ring
B. Adenine, DNA/RNA C. Guanine, DNA/RNA |
|
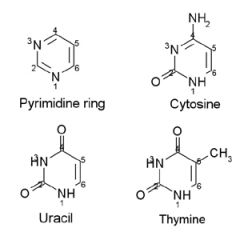
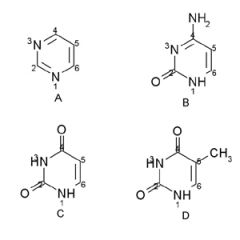
Cytosine, Uracil, and Thymine have ___ ring stuctures.
DNA contains: RNA contains: |
Cytosine, Uracil, and Thymine have Pyrimidine ring stuctures.
DNA contains: cytosine & thymine RNA contains: cytosine & uracil |
|
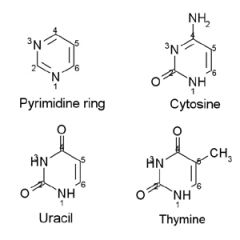
Name the structures and if they are found in DNA and/or RNA.
|
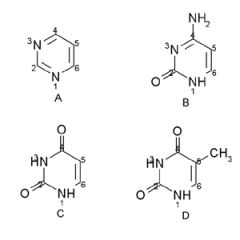
A. Pyrimidine ring
B. Cytosine DNA/RNA C. Uracil RNA D. Thymine DNA |
|
|
1. ___ are compounds in which purine or pyrimidine base is covalently linked to the sugar at carbon # ___.
2. In RNA the sugar is ___ making ___. 3. In DNA the ___ is ___ at the ___ carbon to afford ___ making ___ |
1. Nucleosides, 1.
2. D-Ribose, ribonucleoside. 3. ribose, reduced, 2, deoxyribose, deoxyribonucleoside. |
|
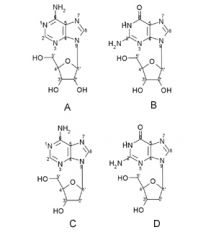
Name these Purine nucleosides:
A. B. C. D. |
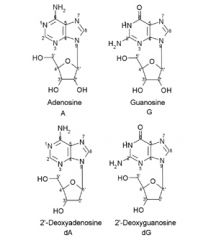
A. Adenosine
B. Guanosine C. 2'-Deoxyadenosine D. 2'-Deoxyguanosine |
|
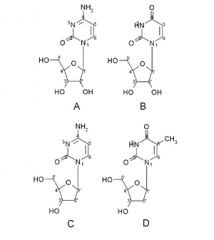
Name these Pyrimidine nucleosides:
A. B. C. D. |
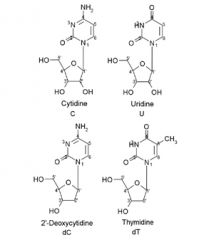
A. Cytidine
B. Uridine C. 2'-Deoxycytidine D. thymidine |
|
|
Nucleotide is the ___ ___ of a nucleoside.
Examples are: AMP: full name ADP: full name ATP: full name |
Nucleotide is the phosphate ester of a nucleoside.
Examples are: AMP: Adenosine 5'-Monophosphate ADP: Adenosine 5'-Diphosphate ATP: Adenosine 5'-Triphosphate |
|
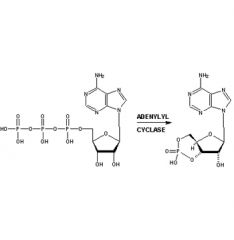
Show mechanism of cAMP formation.
|
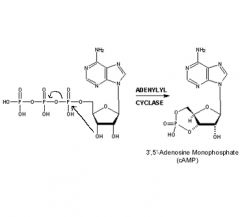
|
|
|
Nucleotides have a wide variety of functions in the body including:
1. 2. 3. 4. 5. |
1. carry activated groups in synthesis of carbohydrates, lipids, and proteins.
2. Structural comp of coenzymes (FAD, NAD+, NADP+, coenzyme A ect) 3. energy sources (ATP, GTP) 4. precursors for DNA/RNA 5. second messengers |
|
|
Their cellular levels are maintained by both ___ ___ synthesis and by ___ ___. Cellular conc of nucleotides are controlled by ___ regulated enzymes. The nucleotide end products of these pathways serve as effectors for key steps.
|
Their cellular levels are maintained by both de novo synthesis and by salvage pathways. Cellular conc of nucleotides are controlled by allosterically regulated enzymes. The nucleotide end products of these pathways serve as effectors for key steps.
|
|
|
Folate Coenzymes are involved in Purine and Pyrimidine Synthesis:
Folic acid must be converted to ___ and then to ___ - which accepts a carbon from serine to become a coenzyme in purine synthesis. The enzyme converting in both steps is ___ and the coenzyme is ___. |
Folic acid must be converted to dihydrofolic acid and then to tetrahydrofolic acid - which accepts a carbon from serine to become a coenzyme in purine synthesis. The enzyme converting in both steps is dihydrofolate reductase and the coenzyme is NADPH.
|
|
|
Methotrexate is a potent inhibitor of ___ ___ in humans.
|
dihydrofoloate reductase
|
|
|
Trimethoprim is a potent inhibitor of ___ ___ in bacteria
|
dihydrofoloate reductase
|
|
|
Folic acid and its coenzymes are ___ ___ into cells.
Once inside they are ___ by an enzyme known as ___ ___. ___ ___ have greater affinity for the folate-dependent enzymes needed for purine and thymidylate (not needed for cytosine & uracil) synthesis, but NOT for ___ ___. |
1. actively transported
2. polyglutamylated, folypolyglutamyl synthetase 3. polyglutamylated folates, dihydrofolate reductase. |
|
|
Interconversion of ___ ___ is necessary for synthesis of purines and thymidine monophosphate.
|
FH4 coenzymes
|
|
|
___ make up the majority of nucleotides found in cells, with ___ being present in the greatest concentration.
|
5'-Nucleotides, ATP
|
|
|
Nucleosides:
In normally functioning cells, ___ ___ predominate. ___ and ___ predominate in hypoxic cells. |
nucleoside 5'-triphosphates
5'-mono, diphosphates |
|
|
de novo Purine biosynthesis:
___ is the common intermediate ("fork in the road") in the synthesis of the other purines. |
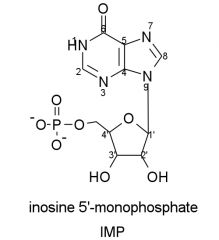
inosine 5'-monophosphate (IMP)
|
|
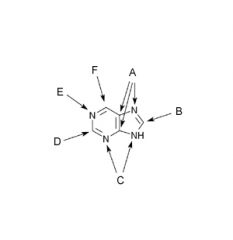
Origin of the atoms in the purine ring.
|
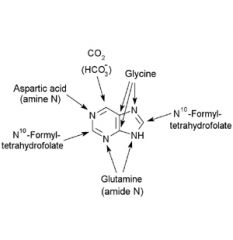
A. Glycine
B. N10-Formyl- tetrahydrofolate C. Glutamine D. N10-Formyl- tetrahydrofolate E. Aspartic acid F. CO2 (HCO3) |
|
|
Ribose 5-phosphate is converted to 5-Phosphoribosyl-1-pyrophosphate (PRPP) by the enzyme ___. This enzyme is activated by ___ and inhibited by ___ ___ ___.
PRPP is also involved in the synthesis of ___ nucleotides and in the ___ ___ rxns in both ___ & ___. |
1. 5-Phosphoribosyl-1-pyrophosphate synthetase
2. Pi (from ATP), purine nucleotide di/triphosphates. 3. pyrimidine, salvage pathway, purines, pyrimidines |
|
|
PRPP stands for ___, it is formed from ___ catalyzed by ___ ___.
|
5-phosphoribosyl-1pyrophosphate, ribose 5-phosphate, PRPP synthetase
|
|
|
From PRPP to IMP (inosine 5'-monophosphate), there are ten steps.
List sites of inhibition here: 3 points of inhibition. |
1. Glutamine PRPP amidotransferase: inhibited by IMP, AMP, GMP, 6-MPRP (6-mercaptopurine ribose phosphate), 6-TGRP (6-thioguanine ribose phosphate).
2. GAR (Glycinamide ribonucleotide) formyltransferase: inhibited by Methotrexate - reduces coenzyme (N10-formyltetrahydrofolate) that transfers the formyl moiety. 3. AICAR (5-aminoimidazole-4-carboxamide ribonucleotide) formyltransferase: Methotrexate again for same reason. |
|
|
Once IMP is synthesized - list the 2 points of end product inhibition and what pathway they are in. This inhibition allows diversion of IMP to synthesis of the purine present in lowest concentration.
|
1. adenylosuccinate synthetase: inhibited by AMP - in the ADP pathway
2. inosate dehydrogenase: inhibited by GMP plus 6-MPRP and 6-TGRP - in the GDP pathway |
|
|
1. 6-MPRP:
2. 6-TGRP: 3. PRPP 4. GAR |
1. 6-mercaptopurine ribose phosphate
2. 6-thioguanine ribose phosphate 3. 5-phosphoribosyl-1pyrophosphate 4. Glycinamide ribonucleotide |
|
|
1. Base ___ nucleoside monophosphate kinases convert nucleoside monophosphates to ___. These enzymes do not discriminate between ___ & ___ sugars in the substrate. Name the 2 kinases:
2. Nucleoside diphosphates are converted to triphosphates by the ___ enzyme nucleoside diphosphate kinase. |
1. specific, diphosphates, ribose, deoxyribose. Adenylate kinase and Guanylate kinase
2. nonspecific (base) |
|
|
Indirect inhibitors of de novo Purine biosynthesis.
1. sulfonamides act as antibacterial agent by inhibiting ___ ___ which is necessary for bacterial synthesis of ___ ___ preventing them from making ___ coenzymes. |
1. dihydropteroate synthetase, dihydrofolic acid, folate
|
|
|
Indirect inhibitors of de novo Purine biosynthesis cont.
Methotrexate inhibits ___ ___ which in needed for synthesis of folate coenzymes in: |
dihydrofolate reductase, humans and animals
|
|
|
Direct inhibitors of de novo Purine Biosynthesis.
1. 6-Mercaptopurine (6-MP) and 6-Thioguanine (6-TG) are metabolically activated by purine salvage pathways to give the ___ ___ analogues. 2. 6-MPRP inhibits: 3. 6-TGRP inhibits: |
1. ribose phosphate
2. amidotransferase, adenylosuccinate synthetase, adenylosuccinate lyase, inosate dehydrogenase 3. amidotransferase, inosate dehydrogenase, guanylate kinase |
|
|
Purine salvage pathways.
1. ___ provides the ribose phosphate group for the 2 enzymes involved. A. ___ catalyzes the rxn with either hypoxanthine or guanine. The products are ___ and ___ respectively. B. ___ catalyzes the rxn with adenine. The product is ___. |
1. 5-Phosphoribosyl-1-pyrophosphate (PRPP)
A. Hypoxanthine-guanine phosphoribosyl transferase (HGPRT), IMP, GMP B. Adenine phosphoribosyl transferase, AMP |
|
|
Purine nucleotide degradation:
1. In humans, the end product of nucleotide degradation is ___. 2. Non-primate mammals oxidize this to ___, which in non-mammal animals is broken further to ___ or ___ in some cases. |
1. uric acid
2. allantoin, urea, ammonia |
|
|
Formation of uric acid:
1. Removal of the amino group from AMP gives ___, amino removal from adenosine gives ___. 2. IMP and GMP are dphosphorylated by ___ to give ___ and ___ respectively. 3. ___ ___ ___ converts inosine and guanosine to ___ and ___ respectively. 4. Deamination of guanine forms ___. 5. ___ ___ oxidizes hypoxanthine to ___ and again to ___ ___, which is secreted in the urine of humans. |
1. IMP, inosine (hypoxanthine riboside)
2. 5'-nucleosidase, inosine, guanosine 3. Purine nucleoside phosphorylase, hypoxanthine, guanine 4. xanthine 5. Xanthine oxidase, xanthine, uric acid |
|
|
1. Xanthine oxidase and xanthine dehydrogenase are two ___ forms of the ___ enzyme.
2. In vivo most mammalian forms of the enzyme exist as the ___. 3. Xanthine oxidase transfers electrons to O2 to form ___ - this is a problem in pathophysiological conditions (ischemia or oxidative stress) in which XD is converted to XO. |
1. interconvertable, same
2. dehydrogenase 3. H2O2 |
|
|
Breakdown of dietary nucleic acids:
1. Pancreatic juice contains ___ & ___ which hydrolyze DNA and RNA to ___. 2. ___ ___ further hydrolyzes these to 3'- and 5'-mononucleotides. ___ then remove the phosphate groups and the nucleosides undergo intestinal absorption. |
1. ribonucleases, deoxyribonucleases, oligonucleotides
2. Pancreatic phosphodiesterase, Nucleotidases |
|
|
Pathology assoc with purine metabolism: Adenosine Deaminase (ADA) Deficiency.
1. ADA catalyzes the rxn of adenosine to ___. 2. Deficiency causes ___ ___ ___. The ___ concentration increases in RBCs, this inhibits ___ ___, thus DNA synthesis is inhibited. *remember - ___ ___ catalyzes ribonucleotide diphosphates to deoxyribonucleotide diphosphates. |
1. inosine
2. severe combine immunodeficiency (SCID) impaired B & T-cell function, dATP, ribonucleotide reductase * ribonucleotide reductase |
|
|
Pathology assoc with purine metabolism: Purine Nucleoside Phosphorylase Deficiency
1. ___ function is impaired, no apparent effect on ___ function. 2. ___ ___ formation is decreased. 3. Purine ___ and ___ concentrations are increased. 4. ___ is the major nucleotide that accumulates in RBCs which stimulates reduction of ADP to dADP which is converted to dATP leading to inhibition of ___ ___. dGTP also inhibits reduction of UDP & CDP. |
1. T-cell, B-cell
2. uric acid 3. nucleoside, nucleotide 4. dGTP, ribonucleotide reductase |
|
|
Pathology assoc with Purine metabolism: Gout
1. Gout is characterized by increased ___ ___ in the blood and urine - AKA ___ & ___ respectively. This leads to attacks of acute ___ ___ ___ due to deposition of uric acid crystals. 2. Primary gout is due to inborn error of metabolism - ie - overproduction of uric acid. 3. Secondary gout is caused by other diseases such as ___(HGPRT) deficiency. 4. Gout may be treated with the ___ ___ inhibitor, allopurinol. |
1. uric acid, hyperuricemia, hyperuricuria, arthritic joint inflammation
3. Hypoxanthine-guanine phosphoribosyltransferase (salvage pathway!) 4. xanthine oxidase |
|
|
Pyrimidine Biosynthesis and Metabolism:
1. Unlike the synthesis of purines (built on top of ribose phosphate), the ___ ___ is built first and attached to the ___ ___ to form the nucleotide. A. ___ (from CO2) and ___ (from hydrolysis of amide of glutamine) plus 2 ATPs give carbamoyl phosphate. B. ___ provides N1 and C4-C6 of the ring. |
1. pyrimidine ring, ribose phosphate
A. Bicarbonate, ammonia B. Aspartate |
|
|
Synthesis of Pyrimidine ring:
1. Carbamoyl phosphate synthetase II is inhibited by ___ and is activated by ___ and ___. Differences btw CPS I and II (chart) |
1. UTP, ATP, PRPP.
|
|
|
The first 3 steps of pyrimidine biosynthesis ctalyzed by ___, ___ ___, and ___, give dihydroorotic acid. These 3 enzymes are all ___ of the ___ polypeptide chain.
|
CPS II, aspartate carbamoyltransferase, dihydroorotase, domains, same
|
|
|
1. Once dihydroorotic acid is formed, it is oxidized to ___ acid by ___ ___.
2. Orotate phosphoribosyltransferase the catalyzes the coupling of PRPP to give orotidine 5'-monophosphate (OMP), decarboxylation by orotate decarboxylase gives UMP. UMP synthatase is a bifunctional enzyme on which ___ ___ ___ & ___ ___ are separate domains on the same ___ ___. |
1. orotic acid, dihydroorotate dehydrogenase
2. orotate phosphoribosyl transferase, orotate decarboxylase, polypeptide chain |
|
|
UMP is changed to UTP using ___ ATP. UTP is used in ___ synthesis.
|
2
|
|
|
Synthesis of CTP from UTP is catalyzed by ___ ___ ___ using ___ as a nitrogen source. CTP is used in ___synthesis, or it can be used to synthesize ___ used in DNA synthesis.
|
Cytidine triphosphate synthetase, Gln (glutamine), RNA, dCTP
|
|
|
UDP is reduced to dUDP by ___ ___. dUDP is loses a phosphate to form Deoxyuridine monphosphate (dUMP). dUMP is methylated by ___ ___ (one carbon transfer) using coenzyme N5,N10- methylene-tetrahydrofolate (FH4), formation of FH4 from FH2 by ___ ___ can be inhibited by ___.
|
ribonucleotide reductase, Thymidylate synthetase, dihydrofolate reductase, methotrexate
|
|
|
Remember, ___ are required for DNA synthesis and are biosynthesized from ribonucleotide diphosphates by the enzyme ___ ___.
|
Deoxyribonucleotides, ribonucleotide reductase
|
|
|
1. Nucleotides are phosphate esters of ___ (1, 2, or 3 phosphate groups attached).
2. Nucleosides are a purine or pyrimidine base attached to a ___ (RNA) or ___ (DNA). |
1. nucleosides
2. D-ribose, D-2'-deoxyribose |
|
|
Give nucleotide functions.
1. 2. 3. 4. |
1. energy source for cellular processes
2. structural components of coenzymes 3. second messengers in signal transduction 4. precursors for DNA and RNA sythesis |
|
|
*DNA is a nucleic acid linked by ___ bonds. DNA has 2 ___ strands. DNA has a ___ and ___ groove.
|
phosphodiester, anti-parallel, major, minor
|
|
|
DNA hydrogen bonds:
1. A purine base pairs with a ___ base. 2. There are ___ H-bonds between adenine and thymine. 3. There are ___ H-bonds between guanine and cytosine |
1. pyrimidine
2. 2 3. 3 |
|
|
The 3D structure of DNA depends mainly on:
1. 2. |
1. hydrogen bond formation
2. base stacking |
|
|
*DNA conformations:
In ___, a right handed spiral is most common. |
B-DNA
Other conformations are A-, H-, Z- |
|
|
1. In a normal (relaxed) DNA molecule, the two DNA strands make a full twist every ___ base pairs.
2. If the molecule has more twists, the strands will coil around eachother causing ___ ___. 3. If a molecule has less twists it is considered to be ___ ___. |
1. 10.4
2. positive supercoiling 3. negatively supercoiled |
|
|
*
___ are enzymes that can monitor and adjust the number of supercoils in DNA |
Topoisomerases
|
|
|
Type ___ topoisomerases can make a break in both DNA strands, ___/___ the DNA, and ___ the breaks.
|
2, wind/unwind, reseal
|
|
|
Type ___ topoisomerases make a break in one of the DNA strands, ___/___ the DNA, and ___ the break.
|
1, wind/unwind, reseal
|
|
|
Topoismoerases play a role in:
1. 2. 3. |
1. DNA replication
2. separation of new DNA during cell proliferation 3. RNA transcription |
|
|
Differences btw prokaryotes vs eukaryotes:
|
Prokaryotes have circular DNA and no nuclear membrane.
Eukaryotes have DNA in nucleus inside nuclear membrane. |
|
|
Prokaryotic and eukaryotic DNA is surrounded by structural proteins called ___.
|
chromatin
|
|
|
*DNA in prokaryotes:
1. The ___ chromosome contains ___ to ___ base pairs that are compacted into 40 to 50 loops of supercoiled DNA - aka ___. 2. Sometimes prokaryotes have plasmids in addition with ___ to ___ basepairs. |
1. circular, 600000 to 5000000, nucleoid
2. 1000 to 10000 |
|
|
*
1. Eukaryotic DNA is very long, humans 3.2 billion base pairs = ___ meters. 2. Eukaryotic chromosomes consists of ___. The DNA structural proteins are ___. |
1. 1-2 meters
2. nucleosomes, histones |
|
|
___ base pairs are wrapped around a core of ___ histone proteins. There are ___ to ___ base pairs in linker DNA.
|
146, 8, 20 to 90
|
|
|
*
1. Mitochondria have their own ___ DNA. 2. Human mitochondrion DNA consists of ___ base-pairs. Each mitochondrion has ___ to ___ copies of this DNA molecule. 3. About ___% of the mitochondrial proteins are encoded by the mitochondrial DNA. |
1. circular
2. 16000, 2 to 10 3. 5 |
|
|
Baltimore classification of viruses.
I. II. II. IV + V. VI + VII. |
I. double stranded DNA viruses
II. single stranded DNA viruses III. double stranded RNA viruses IV + V. single stranded RNA viruses VI + VII. retroviruses |
|
|
*The genome of a DNA virus is a single molecule (___ to ___ base pairs), which is ___ or ___.
I. Double stranded DNA viruses a. b. II. Single stranded DNA viruses a. b. |
The genome of a DNA virus is a single molecule (5000 to 360000 base pairs), which is linear or circular.
I. Double stranded DNA viruses a. Adenoviruses b. Herpes viruses II. Single stranded DNA viruses a. Adeno-associated viruses b. Bacteriophages |
|
|
___ (class I) and ___ (adeno-associated) are used in gene therapy. Both are taken up in the cell via ___. The DNA is transferred to the nucleus via ___ ___. The DNA is ___ incorporated in the cellular DNA.
|
ADENOVIRUSES (class I) and ADENO-ASSOCIATED (class II) are used in gene therapy. Both are taken up in the cell via ENDOSOMES. The DNA is transferred to the nucleus via NUCLEAR PORES. The DNA is NOT incorporated in the cellular DNA.
|
|
|
DNA has a ___ and a ___ groove.
|
major, minor
|
|
|
1. DNA replication is ___, the 2 parental strands are separated and each is used as a template for synthesis of a new strand.
2. The two new DNA molecules each contain one ___ and one ___ strand. |
1. semiconservative
2. parental, new |
|
|
*Three stages of DNA replication.
1. 2. 3. |
1. Initiation - DNA helix is opened up and the DNA replication proteins are positioned.
2. Elongation - sythesis of the new DNA strands 3. Termination - DNA sythesis is stopped |
|
|
Replication Initiation:
Replication begins at the ___ ___ ___, from here 2 ___ ___ are formed. |
origin of replication, replication forks.
|
|
|
Replication Initiation:
1. In each replication fork, the 2 DNA strands are separated by ___. 2. ___ ___ ___ ___ (SSB) bind and hold the separated strands apart. |
1. helicase
2. single-stranded DNA binding proteins |
|
|
Replication Initiation:
___ ___ unwinds the DNA while it is being separated. |
Topoisomerase I
|
|
|
Replication Initiation:
The anti-cancer drug ___ blocks the action of human ___ ___, inhibiting the unwinding of DNA during replication. |
topotecan, topoisomerase I
|
|
|
*Replication Elongation:
1. ___ ___ follow the nucleotide sequence of the template strand in the ___ to ___ direction and synthesize the new strand ___ to ___. 2. The substrates of DNA polymerases are ___ ___ (dATP, dGTP, dTTP, dCTP) |
1. DNA polymerases, 3' to 5', 5' to 3'
2. deoxyribonucleoside triphosphates |
|
|
Replication Elongation:
A phosphodiester bond is formed between the ___ OH group of the last deoxynuleotide in the DNA and the innermost ___ atom of the new deoxynucleoside triphosphate. |
3', phosphor
|
|
|
*DNA polymerases make a mistake only once every ___ to ___ deoxynucleotides. If a mismatch does occur, ___ ___ can remove the mispaired deoxynucleotides - AKA ___.
|
10000 to 1000000, DNA polymerases, proofreading
|
|
|
DNA polymerase proofreading = ___ to ___ exonuclease activity.
|
3' to 5'
|
|
|
1. DNA polymerases can only add deoxynulceotides to an ___ ___ - they need a free ___ group to start adding, this is provided by a primer which is an ___ molecule.
2. The RNA primer is synthesized by ___, which is an ___ ___. |
1. existing strand, 3'-hydroxy, RNA
2. primase, RNA polymerase |
|
|
*
1. DNA sythesis occurs continuously from 5' to 3' on the ___ ___. 2. DNA sythesis occurs in fragments - AKA - ___ ___ in the ___ ___. |
1. leading strand
2. Okazaki fragments, lagging strand |
|
|
1. In eukaryotes, Okazaki strands are ___ to ___ nucleotides long.
2. In prokaryotes, Okazaki fragments are ___ to ___ nucleotides long. 3. The reason for the difference is the ___ from ___ in eukaryotes means fragments are smaller. |
1. 100 to 200
2. 1000 to 2000 3. unbinding, histones |
|
|
Each Okazaki fragment, needed to duplicate the lagging strand of the DNA molecule, starts off with an ___ ___.
|
RNA primer
|
|
|
RNA Primer:
1. Once DNA synthesis is completed, the RNA primer is removed by ___ ___. 2. ___ ___ fills in the gap where the RNA primer used to be. 3. ___ ___ seals the last two nucleotides together. |
1. RNA hybridase
2. DNA polymerase 3. DNA ligase |
|
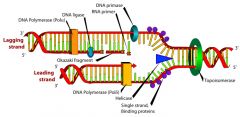
DNA replication summarized:
|
DNA replication summarized:
|
|
|
DNA replication in Prokaryotes:
___ ___ unlinks the two new chromosomes. |
DNA gyrase (topoisomerase II)
|
|
|
DNA polymerases in Prokaryotes:
1. Pol I works as ___ ___ and sythesizes DNA within the gap where the ___ ___ used to be. 2. Pol II is a slow working ___ ___ that comes into action when other ___ can no longer work due to ___ ___. 3. Pol III is the ___ polymerase for DNA synthesis. |
1. RNA hybridase, RNA primer
2. DNA polymerase, polymerases, DNA damage 3. main |
|
|
Some anti-biotics work by inhibition of bacterial DNA synthesis.
1. quinolones inhibit ___ ___. 2. sulfonamides inhibit synthesis of ___, necessary for purine and pyrimidine synthesis. 3. These are both ___ rather than ___. |
1. DNA gyrase
2. folate 3. bacteriostatic, bactericidal |
|
|
*Eukaryotic DNA has ___ origin(s) of replication - it starts at all ___ at the same time.
Prokaryotes have ___ orgin(s). |
multiple, origins, one
|
|
|
DNA polymerases in Eukaryotes:
Pol alpha and Pol delta are involved in DNA replication. 1. Pol alpha forms a comlex with ___ and elongates the ___ with the first ___ to ___ deoxynucleotides. 2. Pol delta is the ___ polymerase for ___ ___. 3. Pol ___, ___, and ___ are involved in DNA repair. 4. Pol ___ replicates mitochondrial DNA. |
1. primase, primer, 15 to 30
2. main, DNA synthesis 3. Beta, delta, e (epsi) 4. gamma 4. |
|
|
DNA polymerase in not capable of replicating the DNA all the way to the end of the chromosome. During each round of replication, some DNA is ___.
|
lost
|
|
|
*
___ are sequences of DNA at the end of eukaryotic DNA that serve as a ___ to prevent ___ of vital genetic information. |
Telomeres, buffers, loss
|
|
|
___ is an enzyme that adds DNA to the end of telomeres. Uses a small ___ molecule as a template.
|
Telomerase, RNA
|
|
|
___ and ___ ___ cells have high telomerase activity. Most cancer cells have ___ telomerase levels.
|
Stem, white blood, high
|
|
|
Telomer is not a ___ structure. The overhanging end forms a loop that is stabilized by telomer binding proteins. ___ are formed from guanosines.
|
straight, G-quadruplexes
|
|
|
Telomeres with G-quadruplexes are no longer accessible for the enzyme ___. Introducing G-quadruplexes in telomeres may inhibit ___ growth.
|
telomerase, cancer
|
|
|
*
1. DNA methylation is found in ___ and ___. 2. It occurs by ___ at the cytosines in CG base-combinations (CpG - side by side, not base pairs), which are often found in clusters (___). 3. ___% of all CpG combinations have a methylated cytosine. In humans this is about ___% of all nucleotides. |
1. prokaryotes, eukaryotes
2. DNA methyltransferase, CpG islands 3. 70, 1 |
|
|
*DNA methylation:
Each cell type in the human body has it's own ___ pattern. This is copied during DNA replication. |
methylation
|
|
|
DNA methylation:
DNA replication is semiconservative. 1. Each ___ strand keeps its methylation pattern. 2. The methylation pattern is copied onto the new DNA strands by ___ ___. |
1. parental
2. DNA methyltransferase |
|
|
*Functions of DNA methylation:
1. ___ ___ is the silencing of one of two alleles of certain genes throught lifetime. |
1. Genomic imprinting
|
|
|
Functions of DNA methylation:
X-chromosome inactivation in females, one of the x chromosomes is silenced ___. |
randomly
|
|
|
*Functions of DNA methylation:
___ ___ is a specialized form of DNA repair that recognizes wrongly placed nucleotides during DNA replication.The mismatch repair machinery is able to recognize the ___ strand. |
mismatch repair, unmethylated
|
|
|
*DNA recombination:
___ is unwanted DNA recombination. |
translocation
|
|
|
*Homologous recombination is found:
1. 2. |
1. DNA repair, one homologous chromosome used as template to repair double strand break in another
2. crossing over during meiosis |
|
|
*Non-Homologous Recombination = ___ ___, requires enzymes that recognize specific nucleotide sequences.
A. Mobile genetic elements 1. Viruses 2. ___ - small segments of that move around in the genome B. formation of antibodies C. Form of DNA damage repair |
site-specific recombination
2. transposons |
|
|
Transposition is the movement of small segements of DNA (transposons) via non-homologous recombination.
1. ___ ___ - the transposon is moved from one site to another. 2. ___ ___ - the transposon is copied and inserted somewhere else in DNA. 3. ___ ___ - copy themselves via RNA intermediate. |
1. simple transposition
2. replicative transposition 3. retrotransposons |
|
|
in prokaryotes, transposons may contain genes for ___ ___.
In eularyotes, transposons do not contain ___ genes. |
antibiotic resistance, functional
|
|
|
Hemophilia, Duchene muscular dystrophy, severe combine immunodeficiency are examples of inherited diseases caused by ___.
|
transposons
|
|
|
Antibody formation:
1. The human genome is not large enough to contain ___ genes for each antibody that is produced. 2. During B-cell development in the bone marrow, antibody gene segments are ___ into a unique combination by ___ recombination. |
1. separate
2. rearranged, non-homologous |
|
|
Antibody Formation cont:
End result of non-homologous recombination and somatic hypermutation is > ___ possible antibodies. Total genes for: 1. Variable chain: 2. Diveristy chain: 3. Joining chain: 4. Constant chain: |
10^10
1. 150 2. 12 3. 4 4. 8 |
|
|
Translocation nomenclature:
In abnormal exchange if genetic material between non-homologous chromosome, t(4;20) signifies: |
genetic material on chromosomes 4 and 20 have been swapped (abnormal)
|
|
|
Chronic myeloid leukemia is caused by a ___ in a hemapoietic stem cellPart of the BCR gene is fused with the ABL gene. The resulting Bcr-Abl protein is a ___ ___ that constantly stimulates cell proliferation.
|
translocation, tyrosine kinase
|
|
|
*Spontaneous Hydrolysis:
1. Spontaneous depurination: mammalian cells loses about ___ - ___ purines a day. 2. Spontaneous depyrimidation: a mammalian cell loses about ___ pyrimidines per day. 3. Spontaneous hydrolysis damage is repaired by ___ ___ ___. |
1. 2000 - 10000
2. 500 3. base excision repair |
|
|
Oxidative stress: Give common reactive species.
1. 2. 3. 4. |
1. superoxide anion (O2-)
2. hydrogen peroxide (H2O2) 3. hydroxy radical 4. hydroxyl ion |
|
|
Reactive oxygen species can be caused by:
1. 2. 3. |
1. byproducts of oxidative metabolism (e transport chain leak)
2. Produced by inflammatory response (neutrophils/macrophages) 3. ionizing radiation |
|
|
*DNA damage from reactive oxygen species can be in the form of (show repair mech for each):
1. 2. 3. |
1. base oxidation - base excision repair
2. single strand breaks - base excision repair 3. double strand breaks - homologous and non-homologous (end joining) recombination |
|
|
Base oxidation (Guanine)
Ex, formation of 8-oxoguanine no longer pairs with cytosine, but mispairs with ___. |
adenine
|
|
|
Many single and double strand breaks are formed by ___ radicals.
|
OH
|
|
|
Ultraviolet radiation:
1. ___ - long wave: 320-400 nm 2. ___ - medium wave: 280-320 nm 3. ___ - short wave: < 280 4. UVA, UVB, and UVC can all damage ___ and enhance ___ aging. 5. UVB and UVC have high energy , due to shorter wavelengths, and can cause ___ damage. |
1. UVA
2. UVB 3. UVC 4. collagen, skin 5. DNA |
|
|
1. Ultraviolet radiation reacts ___ with DNA (unlike ionizing which forms reactive oxygen species).
2. Main DNA damage is ___ formation between nucleotides that distort the DNA ___. |
1. directly
2. dimer, helix |
|
|
Chemical DNA damage:
1. ___ agents add alkyl groups (methyl or ethyl) to DNA bases. For example, O6 methylated guanine mispairs with ___ during DNA replication 2. ___ agents wedge between bases in the DNA strands. Chemotherapeutic agent cisplatin forms DNA crosslinks that make replication impossible. |
1. alkylating, thymine
2. intercalating |
|
|
Repair on a single strand:
1. Spontaneous hydrolysis, base oxidation, and single strand breaks can be repair via ___ ___ ___. 2. UV damage, chemical adducts are repairs by ___ ___ ___. |
1. base excision repair
2. nucleotide excision repair |
|
|
Base excision repair steps:
1. 2. 3. 4. |
1. DNA glycosylase removes bad base from the ribose backbone.
2. 5' endonuclease cuts at 5' prime end of ribose backbone where base was 3. DNA polymerase Beta replaces with proper base 4. DNA ligase seals the ribose backbone gap |
|
|
Nucleotide Excision repair:
1. 2. 3. |
1. a series of nucleotides (ribose backbone and all) containing the UV or chemical adduct damaged is removed
2. DNA polymerase delta or epsi synthesizes the gap from 5' - 3' 3. DNA ligase seals the gap |
|
|
Nucleotide excision repair:
1. Transcription coupled repair focuses on genes that are being transcribed. ___ proteins recognize gene expression proteins that are unable to proceed because of DNA damage. 2. Global genome repair covers the whole genome. ___ proteins recognize damage and locally unwind DNA. |
1. CS (Cockayne Syndrome)
2. XP (Xeroderma Pigmentosum) |
|
|
*Double strand break (from oxidative stress) repair:
1. occurs by ___ ___, two types of this that occur are ___ ___ and ___ ___ ___. |
1. DNA recombination, homologous recombination, non-homologous end joining
|
|
|
Homologous recombination:
1. Mutations in genes coding for proteins that are involved in homologous recombination such as ___, ___, ___ are strongly correlated with ___. |
ATM, BRCA1, BRCA2, cancer
|
|
|
*Repair of UV damage can be repaired two ways.
1. 2. |
1. nucleotide excision repair
2. Photoreactivation Photolyase - binds to UV damage and reverses bond linking two bases (dimer) |
|
|
*
1. Eukaryotic cell cycle lasts ___ to ___ hours. 2. ___ consists of G1 + S + G2 + G0 phases. 3. ___ cells are in G0 undergoing repair. 4. ___ cells undergo cell death |
1. 12 to 24
2. Interphase 3. Quiescent 4. Senescent |
|
|
In the G1 phase:
1. 2. |
1. Preparation for DNA duplication. Cell grows and duplicates important cell organelles.
2. G1 checkpoint will stop progression into S-phase if cell has too much DNA damage |
|
|
In the S-phase:
1. |
1. Cell duplicates its DNA, each chromosome has 2 identicle chromatids bound by a centromere.
|
|
|
In the G2 phase:
1. 2. |
1. Preparation for mitosis - cell also continues to grow
2. G2 checkpoint will prevent progression into M-phase if too much DNA damage |
|
|
*In the M-phase:
1. |
1. The cell separates its chromosomes (mitosis) and its cytoplasm (cytokinesis) to form 2 daughter cells.
|
|
|
*The stages of mitosis are:
1. 2. 3. 4. |
1. prophase - chromosomes condense (topoisomerases involed)
2. Metaphase - chromosomes line up at the metaphase plate 3. Anaphase - chromosomes are pulled towards poles of cell 4. Telophase - new nuclear membrane formed for each daughter cell |
|
|
Prophase:
Chromosomes condense. 1. The ___ start to form ___ ___ made from protein ___. 2. The cetrosomes move to ___ ___ of the cell |
1. centrosomes, mitotic spindles, tubulin
2. opposite sides |
|
|
Prometaphase:
The centrosomes arrive at opposite sides of the cell. 1. Mitotic spindles now cover the nucleus and connect to the ___ of the chromosomes. 2. The nuclear membrane ___. |
1. centromeres
2. disappears |
|
|
*Metaphase:
Chromosomes line up at the metaphase plate. 1. Metaphase cells are used to prepare a ___. 2. Humans have ___ chromosomes - ___ pairs. |
1. karyotype
2. 46, 23 (diploid) |
|
|
Anaphase:
1. Sister chromatids ___. 2. Mitotic spindles ___ and pull the chromosomes towards the ___. |
1. break
2. shorten, centrosomes |
|
|
Telophase:
1. |
1. A new nuclear envelope forms around the 2 new nuclei and the chromosomes unfold.
|
|
|
*Cell cycle regulation:
1. Occurs by ___ ___. 2. A Cdk is active when it is bound to its specific ___. 3. An activated Cdk activates other proteins that promote the ___ ___. 4. Each phase of the cell cycle has its own ___ and ___. |
1. cyclin-dependent kinases (Cdk)
2. cyclin 3. cell cycle 4. cyclins, Cdk's |
|
|
Cell cycle regulation:
The G1 and G2 checkpoints can inhibit ___, preventing progression into next phase. |
cdk's
|
|
|
*___ ___ are proteins located in the extracellular space that determine cell survival and cell proliferation after binding to their receptor on the surface of a target cell.
|
Growth factors
|
|
|
The effects of growth factors depend on their ___ in the extracellular space.
1. ___ = cell death 2. ___ = cell survival 3. ___ = promotion of cell cycle (S-phase) |
Concentration
1. no growth factor 2. low concentration 3. high concentration |
|
|
1. Vascular endothelial growth factor:
- produced by many cell types - induces proliferation of endothelial cells and formation of ___ ___ ___. 2. Platelet derived growth factor - produced in many cell types and in platelets - induces proliferation of many cell types including ___ cells (wound healing) |
1. new blood vessels
2. epithelial |
|
|
Meiosis:
Normal tissue cells are ___. Gametes are ___ and are formed by meiosis. |
diploid (2n), haploid (n)
|
|
|
*Meiosis stages:
1. 2. 3. |
1. replication of homologous pairs of chromosomes
2. meiosis I - replicated homologous pairs separated, these daughter cells are 1n (sister chromatids remain intact) 3. meiosis II - sister chromatids split and haploid gametes formed |
|
|
DNA recombination: crossing over occurs in ___, when homologous chromosomes start to form pairs.
|
prophase I (in meiosis I)
|
|
|
Properties of cancer cells:
Normal skin: - organized tissue - cells communicate Skin carcinoma: - cells divide ___ - no ___ |
uncontrollably, organization/communication
|
|
|
Mutations are caused by:
1. 2. 3. |
1. Viral DNA that becomes incorporated in the genome can act as an oncogene
- Epstein-Barr virus (Burkitt's Lymphoma) - Hepatitis B virus (liver cancer) 2. inherited mutations 3. incomplete/incorrect DNA damage repair |
|
|
*Cancer genes are genes involved in normal cellular processes such as ___ ___ and ___ ___, which upon mutation contribute to cancer formation.
2 types: 1. 2. |
cell cycle, cell death
1. tumor suppressor genes 2. proto-oncogenes |
|
|
Tumor Suppressor Genes:
1. Normal genes that suppress cell ___ or promote ___. 2. A mutation in a tumor suppressor gene may lead to ___ of this gene, which ___ tumor formation. |
1. division, apoptosis
2. inactivation, promotes |
|
|
*Functions of P53:
1. Normal conditions, p53 is ___ 2. DNA damage will lead to ___ and ___ of p53. a. This inhibits ___ ___ ___ leading to ___ ___ ___. b. it also causes release of ___ from ___ leading to ___. |
Functions of P53:
1. Normal conditions, p53 is degraded 2. DNA damage will lead to phosphorylation and activation of p53. a. This inhibits cyclin dependent kinases leading to cell cycle arrest. b. it also causes release of cytochrome-C from mitochondria leading to apoptosis. |
|
|
1. Normal active p53 holds the cell cycle at ___ or induces ___.
2. > ___% of human tumors have a mutation that leads to the ___ or ___ of p53. |
1. G1/S, apoptosis
2. 50, loss, inactivation |
|
|
Functions of the Retinoblastoma Protein (Rb):
1. Regulates the cell cycle by inhibiting a transcription factor that induces ___ ___ progression. 2. Cyclin-dependent kinases 2 and 4 ___ Rb by phosphorylation. |
1. cell cycle
2. inactivate |
|
|
*
1. The Retinoblastoma (Rb) gene is a common ___ ___ gene. 2. The normal Rb protein inhibits the ___ ___. 3. >___% of human tumors have a mutation that leads to the ___ or ___ of Rb. 4. Rb gets its name from ___, a cancer caused by loss of the Rb gene. |
1. tumor suppressor
2. cell cycle 3. 70, loss, inactivation 4. retinoblastoma |
|
|
Human Papillomavirus (HPV) is a ___ stranded virus.
1. To replicate it needs ___ cells. 2. HPV proteins inhibit ___ and ___ protein to induce ___ ___. |
double stranded
1. proliferating 2. p53, Rb, cell proliferation - both p53 and Rb are tumor suppressor genes. |
|
|
Proto-oncogenes are normal genes that often regulate division or apoptosis, but over-promote division or inhibit apoptosis upon mutation.
1. After mutation, proto-oncogenes become ___. |
1. oncogenes
|
|
|
*
1. Ras is a common ___. 2. Growth factors act on surface of cell on ___ ___ ___ which in turn activate Ras which then activates a ___ ___ ___ that activates a transciption factor to cause synthesis of ___ ___. |
1. proto-oncogene
2. receptor tyrosine kinase, protein kinase cascade, cyclin D1 (this leads to cell cycle progression) |
|
|
Ras is a common Proto-oncogene.
About ___% of human cancers have a mutation in the Ras gene that leads to activation of Ras. |
30
|
|
|
Growth factors and their receptors can be ___.
Both ___ ___ and ___ of growth factors and their receptors are found in tumors. - Vascular endothelial growth factor (VEGF) - Platelet derived growth factor (PDGF) - Fibroblast growth factor (FGF) |
proto-oncogenes, activating mutations, overexpression
|
|
|
*Summary of common cancer genes:
Tumor suppressor genes: 1. 2. Proto-oncogenes: 3. 4. 5. |
Tumor suppressor genes
1. p53 2. Retinoblastoma (Rb) Proto-oncogenes 3. Ras 4. growth factors 5. growth factor receptors. |
|
|
Chronic Myeloid Leukemia:
___ is a molecule that binds to Bcr-Abl in its kinase domain and blocks its activity. It is the first drug that directly blocks a protein known to cause cancer. |
Gleevec
|
|
|
Characteristics of RNA
1. contains these ribonucleotides: 2. the sugar in the backbone is ___ instead of ___ (DNA) 3. Contains ___ instead of ___ (DNA) 4. Cellular RNA is usually ___ ___. 5. Viral DNA can be ___ and ___. 6. RNA has complex and diverse 3D structures. |
1. contains these ribonucleotides: Cytidine, Adenine, Uracil, Guanine
2. the sugar in the backbone is ribose instead of deoxyribose (DNA) 3. Contains uracil instead of thymine (DNA) 4. Cellular RNA is usually single stranded. 5. Viral RNA can be single-stranded and double-stranded. 6. RNA has complex and diverse 3D structures. |
|
|
Ribonucleases (RNases)
Enzymes that degrade RNA 1. ___ cleave phosphodiester bond within an RNA molecule. 2. ___ cleave nucleotide from the end of an RNA molecule. |
1. endoribonucleases
2. exoribonucleases |
|
|
Roles of ribonucleases:
1. Regulation of intracellular levels of ___. 2. ___ of RNA molecules 3. Protection against RNA ___. *RNA molecules have ___ stability |
1. RNA
2. Processing 3. viruses * low - unlike DNA which is stable |
|
|
Coding or non-coding RNA:
1. mRNA 2. rRNA 3. tRNA 4. catalytic RNA |
1. mRNA = coding
2. rRNA = non-coding 3. tRNA = non-coding 4. catalytic RNA = non-coding |
|
|
___% of cellular RNA is mRNA
|
5
|
|
|
Eukaryotic mRNA has:
1. 5' cap: a modified ___ nucleotide that helps the initiation of protein sythesis from the mRNA molecule. 2. 3' poly-A tail: up to ___ ___ nucleotides that protects the mRNA from degradation and helps its transport out of the nucleus. |
1. guanine
2. 200 adenine |
|
|
5' cap on mRNA is made from a guanosine methylated at the ___ position which is then attached to the mRNA ___ by a ___ link.
|
N7, backwards (5' to 5'), triphosphate
|
|
|
tRNA = ___% of cellular RNA
|
10
|
|
|
1. Transfer RNA is ___ to ___ nucleotides long.
2. The amino acid is bound to the ___ end of tRNA by ___ ___. 3. ___ is a tRNA molecule with it's amino acid bound to it. |
1. 65-110
2. 3', aminoacyl-tRNA synthetases 3. aminoacyl-tRNA |
|
|
Eukaryotic rRNA molecules are synthesized in 4 sizes:
1. 2. 3. 4. |
1. 28S
2. 18S 3. 5.8S 4. 5S |
|
|
1. About ___% of the mitochondrial proteins are prodiced by the mitchondira itself.
2. Mitochondrial RNA = about ___% of total RNA in the cell. |
1. 5
2. 5 |
|
|
1. Catalytic RNA can be an ___ (catalytic or ribozyme)
2. Ribosomal RNA catalyzes the sythesis of ___. 3. Ribonuclease P is an RNA molecule that cleaves precursor RNA to form ___ tRNA. |
1. enzyme
2. proteins 3. mature |
|
|
RNA primer for DNA synthesis is synthesized by ___.
|
primase.
|
|
|
RNA-induced silencing complex is activated by ___ in order to regulate levels of mRNA in cells.
|
MicroRNA
|
|
|
1. silencing RNA or Small interfering RNA (siRNA) is similar to ___ but only found in ___ and lower ___.
2. When introduced to cells can block and break down mRNAs by binding to the ___ complex - making it useful for research and clinical use. |
1. MicroRNA, plants, eukaryotes
2. RISC (RNA induced silencing complex) |
|
|
RNA molecules in the clinic:
1. ribozymes and siRNA will bind specific mRNA molecules --> inhibit mRNA translation and induce mRNA degradation. Problems with this approach are: 1. 2. 3. |
1. toxicity
2. low stability 3. do not naturally enter cell |
|
|
RNA molecules in the clinic:
Stability can be improved with chemical modifications. 1. methyl groups on the 2' oxygen of ribose protect against ___. 2. sulfur replacement of the 3' oxygen protects against ___. |
1. endonucleases
2. exonucleases |
|
|
RNA molecules in the Clinic:
1. ___ can be used to carry the siRNA and fuse with the cell membrane. 2. A ___ ___ can be used to inject the siRNA into the cell. |
1. liposome
2. viral vector |
|
|
Common viral vectors:
1. ___ - replicate their genome via a DNA intermediate (reverse transcription). The DNA gets incorporated into the cellular DNA. 2. ___ - double-stranded DNA viruses. The DNA does not get incorporated into the cellular DNA. Daughter cells will not carry the new gene. 3. ___ - single-stranded DNA virus, not know to cause disease. |
1. Retroviruses
2. Adenoviruses 3. Adeno-associated viruses |
|
|
Small interfering RNA (siRNA) is similar to microRNA but:
|
siRNA is found in plants and lower eukaryotes and is sythesized in the lab for research and clinical use. Both bind to mRNA molecules and lead to cleavage (degradation).
|
|
|
Transcription in eukaryotes:
1. ___ - DNA that is actively transcribed is found in a loose form of chromatin 2. ___ - inactive parts of DNA (highly condensed) 3. ___ - DNA that is used to synthesize rRNA (ribosomes) molecules |
1. euchromatin
2. heterochromatin 3. nucleolus |
|
|
RNA polymerases: enzymes that sythesize RNA
1. RNA polymerase I ---> ___ 2. RNA polymerase II ---> ___ 3. RNA polymerase III ---> ___ * most regulation of expression occurs at ___ |
1. RNA polymerase I ---> rRNA
2. RNA polymerase II ---> mRNA 3. RNA polymerase III ---> tRNA * RNA polymerase II |
|
|
Transcription of RNA from DNA:
The template strand is used as a template to synthesize the RNA. 1. The other strand in the DNA is the ___ strand. It has the same sequence as the ___ molecule. It is used to report the sequence of a gene |
1. coding, RNA
|
|
|
1. Initiation: ___ ___ is properly positioned on the DNA of the gene that needs to be transcribed.
2. Elongation - ___ ___ transcribes the DNA sequence of the gene into an RNA molecule. 3. Termination - ___ ___ is released from the DNA |
1. RNA polymerase
2. RNA polymerase 3. RNA polymerase |
|
|
Transcription initiation:
1. The ___ ___ factors bind to consensus sequences in the ___. 2. consensus sequences are located ___ of the gene. They are not translated into RNA 3. Consensus sequences are ___ ___, ___ ___ and ___ ___. |
1. general transcription, promoter (region containing CAAT, GC, and TATA boxes_
2. upstream 3. CAAT box, GC box, TATA box |
|
|
Transcription initiation:
A. The transcription complex is formed. The general transcription factor TFIID binds to the ___ ___ in the promoter region. B. Then other general transcription factors attach. |
A. TATA box
|
|
|
Transcription initiation:
C. The general transcription factors form a complex with ___ ___ called the ___ ___ ___. D. RNA polymerase is now ready to leave the initiation complex and start RNA sythesis. |
C. RNA polymerase, transcription initiation complex
|
|
|
Transcription elongation:
1. RNA polymerase locally separates the 2 strands of DNA to form an opening +- 10 mucleotides long. 2. DNA unwinding is facilitated by ____ 3. RNA polymerase transcribes the template strand of the DNA into an RNA molecule. 4. The RNA molecule is synthesized from ___ to ___. |
2. topoisomerases
4. 5' to 3' |
|
|
Transcription termination:
1. not well understood in eukaryotes 2. RNA polymerase can continue for ___ to ___ nucleotides beyond the end of the gene before it releases from the mRNA 3. The ___ RNA is the processed to give correct length. |
2. 1000 to 2000
3. precursor |
|
|
All mRNA processing occurs in:
|
the nucleus
|
|
|
Adding the mRNA 5' cap:
Give enzymes and process: 1. 2. 3. 4. |
1. phosphatase cleave the 5' terminal end's outer most phosphate
2. Guanylyl transferase attaches a GTP's 5' end inner most phosphate to the 2 phosphates on the 5' end of the mRNA - forming a triphosphate 5' to 5' bond. 3. Methyl transferase methylates N7 on the former GTP 4. methyl transferase methylates 2' oxygen on last couple of nucleotides |
|
|
Adding the Poly(A) tail:
Occurs at the ___ ___ (AAUAAA) in the mRNA. Hundreds of adenosines can be added |
polyadenylation signal
|
|
|
Splicing:
Genes contain ___ (coding sequences) and ___ (intervening sequences). |
exons, introns
|
|
|
Alternative splicing:
1. The human genome has +-___ protein-coding genes. 2. ___ to ___ different proteins are produced. * This can be explained by ___ ___. |
1. 36,000
2. 80,000 to 100,000 * alternative splicing |
|
|
List the 4 difference between DNA and RNA sythesis:
1. 2. 3. 4. |
1. one makes DNA, the other RNA
2. RNA uses RNA polymerase, DNA uses ....duh! 3. DNA needs primer, RNA synth does not. 4. DNA is proofread, RNA is not 4. |
|
|
___ ___ - process by which DNA molecule is formed from an RNA molecule. Performed by ___ ___.
*example: ___ |
Reverse transcription, reverse transcriptases
* Telomerase |
|
|
The genome of RNA viruses is an ___ molecule.
1. most have ___ stranded RNA 2. Some have double stranded RNA such as ___. 3. They repliate their genome by using a viral ___ ___. |
RNA
1. single 2. reoviruses 3. RNA polymerase |
|
|
Retroviruses:
1. RNA viruses that replicate their genome via a ___ intermediate formed via viral ___ ___. 2. The viral DNA is incorporated into the cellular DNA via non-homologous recombination 3. Lack of proof reading in reverse transcriptases leads to rapid changes in viral genome. |
1. DNA, reverse transcriptase
|
|
|
Regulation of gene expression:
1. ___ ___ are always expressed at a constant level. Most are involved in maintenance of the cell. Ex: ___ ___ 2. Most genes are expressed only when needed. 3. Some are cell specific. Ex: insulin gene only expresed in beta cells of pancrease |
1. housekeeping genes, ribosome genes
|
|
|
1. Regulation of mRNA sythesis is about ___% of all gene expression regulation.
2. Regulation of mRNA stability is ___% of all gene expression regulation. |
1. 60%
2. 40% |
|
|
1. Nuclear receptors are a class of transcription factors that are activated by ___ and ___ ___.
2. These then bind to ___ ___ ___ in the DNA to induce gene transcription. 3. INactive nuclear receptors can be located in the ___ or the ___ of the cell. |
1. steroid, thyroid hormones
2. hormone response elements 3. cytoplasm, nucleus |
|
|
1. Activators are proteins that enhance gene expression but do not ___ gene expression by themselves.
2. They bind to an ___ ___ in the DNA. 3. These can be close to or far away (___ of base pairs away) from the promoter. 4. They can be ___ or ___ from the promoter. 5. May interact with transcription factors and RNA polymerase by ___ the DNA |
1. induce
2. enhancer element 3. 1000's 4. upstream, downstream 5. bending |
|
|
1. Repressor are ___ of activators
2. they bind to a ___ in the DNA, which can overlap with: a. b. |
1. counterparts
2. silencer a. activator binding site b. transcription start site |
|
|
Repressors in disease:
A double mutation in the Wilms Tumor (WT1) gene leads to kidney tumors at an early age. 1. The WT1 protein is a ___ ___ that inhibits the expression of EGR-1 (early growth response factor-1). 2. Lack of inhibition of EGR-1 leads to ___ ___. *WT1 is considered a ___ ___ gene. |
1. transcription repressor
2. uncontrolled growth *tumor suppressor |
|
|
DNA methylation occurs at ___ in ___ ___. Then ___ ___ bind to these sites (gene inactivation) and the transcription initiation complex can no longer be formed.
|
cytosines, CpG islands, Methyl-CpG-binding proteins
|
|
|
DNA methylation in cancer
Hypermethylation of ___ regions of genes is common in cancer cells. |
promoter
|
|
|
Histone acetylation:
___ ___ adds acetyl groups to ___ amino acids in the histone protein, which takes away their net ___ ___. This ___ the interaction btw histones and the DNA. |
Histone acetyltransferase, lysine, positive charge, losens
|
|
|
Acetylation of histones in cancer:
1. ___ of histone acetylation is found in many tumors. 2. This leads to ___ ___ and reduced ___ ___. |
1. loss
2. gene silencing, DNA repair |
|
|
___ ___ contain proteins that degrade mRNA when a cell is under stress.
|
stress granules
|
|
|
P-bodies are similar to stress granules but contain ___. The ___ ___ is thought to bring the mRNA into the P-bodies
|
microRNA, RISC complex (RNA induced silencing)
|

