![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
191 Cards in this Set
- Front
- Back
|
autotroph |
produces own food |
|
|
heterotroph |
consume external energy sources |
|
|
CHNOPS |
six most common elements in organic matter Carbon, Hydrogen, Nitrogen, Oxygen, Phosphate, Sulfur |
|
|
lineage |
changes in the program of an organism over time; allows evolution |
|
|
Linnaeus |
Swedish biologist established binomial nomenclature |
|
|
variability |
enables favorability to traits (eg hair length) |
|
|
heritability |
traits passed through genetics (eg height) |
|
|
fitness |
ability to produce offspring relative to others in the population |
|
|
biological imperative |
thrust your genes into the next generations |
|
|
Haekel |
added protista to plantae and animalia in 1866 |
|
|
protista |
colonial single-celled organisms (eg algae) |
|
|
Robert Whittaker |
added monera (bacteria) and fungi in 1969 |
|
|
Woese |
"scarred revolutionary" revolutionized taxonomy by organizing organism by biochemical similarity in ribosomal RNA |
|
|
domains |
of life: bacteria, archae, eukarya |
|
|
isotopes |
"equal places," are variants of an element by # neutrons |
|
|
atomic weight |
average of mass #s of naturally occuring isotopes by abundance %s |
|
|
radioactive isotope |
neutrons change into protons, changing the element (eg 14C --> 14N), helpful in fossil dating |
|
|
dalton |
unit of mass measuring atoms, equivalent to a proton or neutron |
|
|
orbital |
region in which up to 2 electrons surround the nucleus |
|
|
electron shell |
levels of orbitals |
|
|
valence shell |
outermost electron shell |
|
|
valence electrons |
electrons in the valence shell |
|
|
valence |
the # of unpaired electrons in an atom's valence shell (eg, oxygen's valence is 2) |
|
|
molecules |
substances held together by covalent bonds |
|
|
covalent bonds |
shared electrons; when attractive forces (electron-proton) overcome repulsive forces (electron-electron or proton-proton) |
|
|
electronegativity |
an atom's ability to attract electrons, affected by # protons and # electron shells; O > N > C = H
|
|
|
nonpolar |
atoms in a covalent bond share valence electrons equally, with equal strength; uncharged |
|
|
polar |
imbalance in covalently bonded atoms' electronegativities; electrons shared unequally, closer to one atom than another; have partial charge |
|
|
partial charge |
∂ , of an atom in a polar covalent bond, due to electrons (and their charges) being more concentrated on one end of the bond, towards the more electronegative atom |
|
|
ionic |
a bond in which electrons transfer completely from one atom to another (forming ions); ions stick together due to opposite, full charges
|
|
|
ion |
fully charged atoms due to extra or missing electrons |
|
|
cation |
positively charged ions (#protons > #electrons) |
|
|
anion |
negatively charged ions (#electrons > #protons) |
|
|
solvent |
dissolves solutes (breaks down covalent/ionic bonds between atoms of a molecule) to make a solution; water is good at this because it's polar |
|
|
hydrogen bonds |
the weak intermolecular attraction between partially charged H's and O's in water, to other H2O molecules or different (polar) substances |
|
|
hydrophilic |
substance that can dissolve in water; polar |
|
|
hydrophobic |
substance that does not dissolve in water; nonpolar |
|
|
hydrophobic reactions |
interactions between hydrophobic substances submerged in water; H2O forms stronger bonds with H2O, then with hydrophilic substances, not easily with hydrophobic substances |
|
|
cohesion |
when water stays together because of its strong hydrogen bonds |
|
|
adhesion |
when water "sticks" to any polar/charged solid surfaces |
|
|
surface tension |
minimizes surface area, acts like an elastic membrane, outputs reistance; eg surface water molecules have stronger bonds with each other, having fewer surrounding H2O molecules to share hydrogen bonds with than submerged water |
|
|
specific heat |
amount of energy required to raise 1 gram of a substance 1 degree Celsius |
|
|
heat of vaporization |
point at which a substance becomes gaseous; related to specific heat |
|
|
chemical reaction |
when atoms rearrange their bonds, either combining or breaking apart substances |
|
|
dissociation reaction |
when atoms of a molecule separate (eg 2H2O --> H3O + OH-) |
|
|
acids |
donate protons (ie Hydrogen atoms) in a chemical reaction
|
|
|
bases |
accept protons (ie Hydrogen atoms) in a chemical reaction |
|
|
mole |
6.022 x 10^23; a named number like "dozen" which converts a certain mass of a substance to its # molecules to make up that amount of mass; 1 H atom x (6.022 x 10^23) = 1 mole of H |
|
|
molecular weight |
sum of atomic weights in a molecule (eg H2O = 1 + 1 + 16 = 18) |
|
|
molarity |
# moles of a substance in a solution per 1 liter |
|
|
pH |
concentration of protons in a solution; a scale of 1-14, where each integer represents a power of 10; affects polarity and therefore the chances for certain chemical reactions to occur |
|
|
buffers |
substances that offset changes in pH due to chemical reactions in a cell/tissue; maintains homeostasis |
|
|
ammonia |
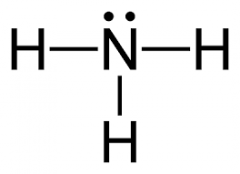
NH3 |
|
|
reactant |
initial molecules in a chemical reaction (left) |
|
|
product |
resulting molecules in a chemical reaction (right) |
|
|
chemical equilibrium |
dynamic but stable state of a substance undergoing spontaneously reversible chemical reactions (<-->) |
|
|
system |
a closed set of interacting elements |
|
|
endothermic |
heat absorbed during a chemical reaction |
|
|
exothermic |
heat released during a chemical reaction |
|
|
energy |
the capacity to do work or supply heat |
|
|
potential energy |
stored energy |
|
|
kinetic energy |
energy of motion |
|
|
chemical energy |
potential energy stored in bonds (distance of shared electrons from a nucleus, highest in nonpolar covalent bonds) |
|
|
thermal energy |
kinetic energy of molecular motion |
|
|
temperature |
how much thermal energy an object's molecules possess |
|
|
heat |
transferred thermal energy between contacting objects of different temperatures |
|
|
first law of thermodynamics |
energy is conserved; cannot be created nor destroyed |
|
|
spontaneous |
chemical reactions which are able to proceed on their own without continuous external influence |
|
|
nonspontaneous |
chemical reactions which require continuous external influence to proceed |
|
|
entropy |
amount of disorder in a system, represented by an italic S |
|
|
second law of thermodynamics |
entropy increases in an isolated system |
|
|
prebiotic soup model |
atmospheric gasses synthesized certain molecules (or came from meteorites), condensed with rain, and accumulated in oceans |
|
|
surface metabolism model |
dissolved gasses formed organic and complex molecules with minerals from deep-sea vents |
|
|
photons |
packets of light energy, can scatter electrons which breaks covalent bonds |
|
|
ozone |
O3, an atmospheric gas which prevents photons from reaching the Earth's surface |
|
|
free radicals |
fragments from photon dismemberment, have unpaired valence electrons |
|
|
catalyst |
a substance that speeds up chemical reactions by providing appropriate chemical environment for reactants to interact effectively; orients reactants
|
|
|
organic molecules |
contain carbon bonded to other elements |
|
|
carbon |
C, atomic # 6, has 4 valence electrons, structures organic molecules |
|
|
functional groups |
amino, carboxyl, carbonyl, hydroxyl, phosphate, sulfhydryl; groups that characterize many substances by providing specific structures and functions |
|
|
amino functional group |
R--NH2, acts a base |
|
|
carboxyl functional group |
R--COOH-, acts as an acid |
|
|
carbonyl functional group |
R--COH or R--CO--R, aldehydes and ketones, help for larger molecules |
|
|
hydroxyl functional group |
R--OH, alcohols, highly polar so helps make compounds more soluble in water, acts as a weak acid |
|
|
phosphate functional group |
O R--OPO- store lots of chemical energy O- |
|
|
sulfhydryl functional group |
R--SH, can form disulfide (S--S) bonds that contribute to protein structure |
|
|
free energy |
amount of energy that is available to do work, represented by an italic G |
|
|
enthalpy |
total energy in a molecule represented by an italic H in formulas; potential energy (heat content) + surrounding's effects (pressure, volume) |
|
|
delta |
Δ, represents change |
|
|
Gibb's free-energy exhange |
determines whether a chemical reaction is spontaneous; ΔG = ΔH - TΔS
|
|
|
exergonic |
spontaneous reactions, increases entropy, ΔG < 0 |
|
|
endergonic |
nonspontaneous reactions, decreases entropy, ΔG > 0 |
|
|
energetic coupling |
endergonic and exergonic reactions facilitate each other with the exchange of free energy |
|
|
redox reactions |
reduction-oxidation reactions; when an atom/molecule loses or gains electrons; OIL RIG |
|
|
electron carrier |
acts as both an electron donor and receptor to relocate H2 |
|
|
ATP |
adenosine triphospate; lots of potential energy (in its 3 phosphate groups, nonpolar, electrons at farthest edges of valence shells) |
|
|
kcal |
kilocalorie; energy it takes to raise 1 kilogram of water 1 degree Celsius |
|
|
substrate |
target molecule that interacts with a catalyst as a reactant |
|
|
phosphorylation |
phosphate group is added to a substrate |
|
|
activated substrate |
phosphorylated substance that acts as an intermediate that allows two other substances to be exergonically reacted |
|
|
amino acids |
20 building block molecules for proteins |
|
|
monomer |
"one part" single molecular subunit (eg amino acid, nucleotide) |
|
|
polymer |
"many parts" several monomers bonded together (eg protein, carbohydrate) |
|
|
polymerization |
linking of monomers into polymers |
|
|
macromolecules |
very large molecules (a type of polymer) made of smaller molecules, nonspontaneously formed |
|
|
condensation reaction |
aka dehydration reaction: water is a product (opposite of hydrolysis), polymerization is this type of chemical reaction |
|
|
peptide bond |
C--N covalent bond that results from a condensation reaction, ACTS as a double bond |
|
|
residue |
a molecule linked by peptide bonds |
|
|
n-terminus |
amino terminus (NH3+), start on left |
|
|
c-terminus |
carboxyl terminus (COO-), end on right |
|
|
ogliopeptide |
a smaller peptide chain of <50 amino acids; called just a "peptide" |
|
|
polypeptide |
50+ amino acids |
|
|
protein |
any chain of amino acid residues, but technically only the complete, functional molecules |
|
|
primary structure |
unique sequence of amino acids in a protein |
|
|
secondary structure |
alpha-helix and beta-pleated sheets; are the folds created by hydrogen bonds between R-groups |
|
|
tertiary structure |
overall shape of a protein, can also include interactions between R-groups and backbone via hydrogen bonds, hydrophobic interactions, van der Waals interactions, disulfide bonding, and ionic bonding |
|
|
quarternary structure |
shape of proteins that contain multiple polypeptides |
|
|
dimer |
"two parts", proteins with 2 polypeptide subunits |
|
|
tetramer |
"four parts", proteins with 4 polypeptide subunits |
|
|
macromolecular machines |
groups of multiple proteins that assemble into one structure to perform a specific function (eg ribosome) |
|
|
denature |
unfolded proteins, done by treating protein with compounds that break disulfide bonds, disrupts function of protein |
|
|
molecular chaperones |
proteins that facilitate folding (usually after heat-shock or for basic activity regulation in enzymes) |
|
|
prion |
alternate "normal" (ie, inactive) protein shapes which make other proteins adopt the dysfunctional shape. fatal disease (eg mad cow disease, spongiform encephalopathies) |
|
|
catalyze |
speed up chemical reactions |
|
|
enzyme |
a protein that acts as a catalyst, large and globular, often end in -ase, flexible and dynamic; stabilize reactions to lower activation energy |
|
|
active site |
where substrates bind to enzymes and where catalysis occurs |
|
|
induced fit |
conformational change of an enzyme when substrate(s) bond at the active site; stabilizes transition state |
|
|
transition state
|
climax of catalysis, when substrate(s) bind through hydrogen bonds and R-groups interact |
|
|
activation energy |
kinetic energy requires to catalyze substrates' reactions |
|
|
initiation |
step 1 of enzyme catalysis: orient reactants non-randomly
|
|
|
transition state facilitation |
step 2 of enzyme catalysis: stabilize reaction by lowering activation energy |
|
|
termination |
step 3 (of 3) of enzyme catalysis: products released from enzyme as they do not fit in active site |
|
|
saturation kinetics |
as substrate levels increase, so does enzyme activity, until substrate levels approach, meet, and exceed enzyme levels, which then function at maximum efficiency |
|
|
cofactors |
a type of enzyme helper made of inorgainic ions (eg Zn2+, Mg2+, Fe2+); reversibly interacts with enzymes |
|
|
coenzymes |
a type of enzyme helper made of organic molecules that reversibly interact with enzymes (eg NADH, FADH2)
|
|
|
prosthetic groups |
a type of enzyme helper made of non amino acids *permanently* attached to enzymes (eg retinal), often attached to active site |
|
|
competitive inhibition |
regulatory molecule is similar in size and shape to substrate, so will occupy active site, thus inhibiting the chemical reaction, and competing with the substrate for enzyme interaction |
|
|
allosteric regulation |
regulatory molecule binds at non active sites, changing the shape of the enzyme and allowing or preventing it to function properly |
|
|
RNA world hypothesis |
suggestion that life began with a nucleic acid, not proteins because they can replicate themselves |
|
|
nucleic acid |
a polymer made of nucleotides |
|
|
nucleotide |
a monomer which contains a phosphate group, a 5-carbon sugar, and a nitrogenous base |
|
|
purines |
adenine & guanine (have 9 atoms) |
|
|
pyrimidines |
cytosine, uracil, and thymine (have 6 atoms) |
|
|
pentoses |
sugars with 5 carbons |
|
|
hexoses |
sugars with 6 carbons |
|
|
ribose problem |
"how did ribose become the dominant sugar in prebiotic soup?"... minerals prefer to bond with ribose (think deep sea vents) |
|
|
phosphodiester bond |
polymerization/ condensation bond between hydroxyl (R--COOH-) and sugar of one nucleotide and phosphate of another (joins 5' and 3' to sugars); forms backbone |
|
|
xray crystallography |
seeing how DNA scatter radiation; used to identity DNA structure |
|
|
antiparallel |
side by side, but in opposite directions |
|
|
double helix |
coiled sugar-phosphate backbone on outside, N bases on inside |
|
|
complementary pairing |
purine-pyrimidine pairs AT/AU/GC |
|
|
template strand |
preexisting DNA sequence |
|
|
complementary strand |
new, opposite sequence |
|
|
hairpin |
most common secondary structure of RNA, stem and loop configuration |
|
|
ribosomes |
catalytic RNA molecules, can catalyze hydrolysis and condensation reactions for phosphodiester linkages (and also peptide bonds) |
|
|
RNA replicase |
enzyme that catalyzes replication of RNA |
|
|
amyloid |
clumps of insoluble fibrous protein
|
|
|
plasma membrane |
aka cell membrane, separates life from nonlife with selective barrier to keep damaging compounds outside, allow entry of needed compounds, and allow more frequent collisions necessary for chemical reactions |
|
|
lipid |
catchall term for carbon-containing compounds found in organisms that are nonpolar and hydrophobic |
|
|
hydrocarbons |
molecules that contain only C and H, nonpolar |
|
|
fatty acid |
lipid with hydrocarbon chain and carboxyl (COOH-) functional group |
|
|
saturated |
a fatty acid with no carbon double bonds (C==C) |
|
|
unsaturated |
a fatty acid with at least one carbon double bond |
|
|
fats |
nonpolar molecules called triaglycerols or triglycerides composed of 3 fatty acids and glycerol |
|
|
oils |
polyunsaturated liquid triaglycerols (ie, fats with a lot of unsaturated fatty acids) |
|
|
glycerol |

a 3-carbon molecule |
|
|
ester linkage |
the connection between a fatty acid and a glycerol molecule |
|
|
steroids |
family of lipids characterized by bulky 4-ring structures |
|
|
phospholipids |
glycerol + phosphate group + 2 hydrocarbon chains (either fatty acids or isoprenoids), plus any small organic molecule |
|
|
amphipathic |
"dual sympathy" a molecule with both hydrophilic and hydrophobic parts (eg phospholipids) |
|
|
isoprenoid |
pure hydrocarbon chain, a type of fatty acid |
|
|
cholesterol |
big bulky rings with isoprenoid tail |
|
|
fluid mosaic model |
proteins operate embedded into membrane |
|
|
channels |
a type of transport protein, lined with pores (passive transport) |
|
|
carrier proteins |
a type of transport protein, aka transporter, binding induces a conformational change (passive transport) |
|
|
pumps |
a type of transport protein which uses energy (ATP) to move ions against electrochemical gradients |
|
|
aquaporins |
channels specific to H2O |
|
|
lipid bilayer |
two sheets of lipid molecules spontaneously aligned |
|
|
micelle |
spontaneously formed ring of fatty acids or simple hydrocarbons, all tails meet in center |
|
|
liposome |
a circular/spherical lipid bilayer often used to transport medicene |
|
|
planar bilayer |
a single bilayer line often used in artificial membrane to cross a gap between a plastic divider between two aqueous solutions |
|
|
permeability |
tendency of a structure to allow a given substance to pass through it, related to fluidity |
|
|
selective permeability |
some substances cross a bilayer much easier than others (small nonpolar, small uncharged polar, large uncharged polar, ions) |
|
|
solutes |
dissolved molecules and ions |
|
|
diffusion |
movement of solutes due to inherent kinetic energy (from high to low concentration) |
|
|
concentration gradient |
difference in concentrations causes net movement of solutes (high to low) |
|
|
osmosis |
diffusion of water (from low solute concentration to high solute concentration through selectively permeable membranes) |
|
|
protocells |
vescile-like structures that harbor nucleic acids |
|
|
hypertonic |
higher concentration than comparison (ie outside > inside, hypertonic to CELL), attracts osmosis |
|
|
hypotonic |
lower concentration than comparison (ie inside > outside, hypotonic to CELL), does not attract osmosis |
|
|
isotonic |
dynamically stable concentrations (ie inside = outside) |

