![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
76 Cards in this Set
- Front
- Back
|
Genome |
The raw material of genetics; the complete sequence of nucleotides of the genetic material |
|
|
Gene |
Nucleotide sequence that can code for a product or set of products depending on factors such as alternative splicing and protein modification Unit of heredity DNA in most cases, but can be RNA in some viruses |
|
|
Trait |
genetically influence characteristic gene is a unit of heredity- sequence of nucleotides that code for a trait (genetically influenced characteristic) |
|
|
Epigenome (simplistic) |
All of the epigenetic changes that affect how a gene is read, and thus, gene expression Keep in mind, the epigenome is not a physical structure, but a fluid process of regulation Histone protein modification and non-coding RNA sequences play a large role Note**Epigenetic mechanisms do not change the genome, they simply adjust what is read and how it's read |
|
|
Central Dogma of gene expression |
DNA transcribed to RNA
RNA translated into amino acids Amino acids to protein *all living organisms use this method to express their genes |
|
|
3 major functions of the genetic code |
coding for necessary products during the lifetime of the cell copying genetic information for the creation of new cells within an organism passing on genetic information to the next generation |
|
|
Chromosomes |
In eukaryotic cells, double-stranded DNA sequences are arranged into chromosomes; chromosomes allow the genome to be compressed and organized Chromosomes consist of compactly wrapped DNA and protein in a hierarchy of organizational levels Sections not in use are wrapped around globular proteins called histones |
|
|
Histones
|
Globular proteins that DNA wraps itself around in chromosomes Histones have a net positive charge at a normal cell pH, thus attracting DNA and assisting in the wrapping process DNA wrapped around histones = not in use |
|
|
Nucleosome |
8 histones wrapped in DNA form a nucleosome |
|
|
Supercoils and solenoids |
8 histones wrapped in DNA form nucleosomes. Nucleosomes wrap into coils called solenoids. Solenoids wrap into supercoils. |
|
|
Chromatin |
The entire DNA complex (including a small amount of RNA) By mass: 33% DNA 66% Protein *Small amount of RNA not all chromatin is equally compact. In fact, the cellular machinery only reads chromatin that is uncoiled |
|
|
Heterochromatin |
Chromatin that is tightly condensed |
|
|
Constitutive heterochromatin |
chromatin that is permanently coiled |
|
|
Euchromatin |
chromatin that is uncoiled and able to be transcribed; only happens during cell division |
|
|
Single Copy DNA |
Nucleotide sequences represented by only one copy of a Nucleotide sequence, and associated with regions of euchromatin that are being actively transcribed |
|
|
Repetitive DNA |
non-coding sequences of DNA (found only in eukaryotes) contain repetitive DNA multiple consecutive copies of the same Nucleotide sequence and remains tightly coiled in regions of heterochromatin
|
|
|
DNA methylation |
Most common example of epigenetic regulation through chemical change addition of an extra methyl group to a particular cytosine nucleotides ; methylation causes DNA to be wound more tightly methylated sections are inaccessible to cellular machinery, and thus, cannot be transcribed expression of genes in these sections are greatly reduced DNA methylation can be inherited! future generations can be more prone to things like asthma, obesity, et cetera |
|
|
non-coding RNA |
sections of RNA that do not code for protein products ; contribute to the regulation of the chemical changes that affect chromatin structure |
|
|
Homologues |
Inside the human somatic cell = 46 double stranded DNA molecules Chromatin associated with each one is wound into a chromosome In human cells, each chromosome possesses a partner that codes for the same traits as itself. two such chromosomes are called - homologues Humans posses 23 homologous pairs of chromosomes |
|
|
Diploid |
any cell that contains homologous pairs of chromosomes |
|
|
Haploid |
any cell that does not contain homologous chromosomes |
|
|
Transcription |
Process by which RNA is manufactured from a DNA template During transcription, an RNA transcript is created (from DNA) Different genes code for different types of RNA mRNA (messenger RNA) is most abundant, which serves as the message that is translated for protein production |
|
|
Transcription in Eukaryotes (where does this happen?) |
Nuclear DNA cannot leave the nucleus Mitochondrial DNA cannot leave the mitochondrial matrix Transcription only happens in these two places |
|
|
Translation |
to produce proteins, transcribed mRNA must undergo the process of translation translation takes the whole nucleotide sequence of the RNA transcript and translates that into the language of amino acids, which are then strung together to form a protein recall that proteins are essential to cell structure, and regulate virtually all cell processes. Given that, this process is important! |
|
|
Purpose of transcription |
create an RNA copy of a DNA template; Transcription is itself a form of regulation of gene expression only the DNA transcribed has an opportunity to be translated into a protein. Without this, all cells would essentially be the same |
|
|
Three main stages of transcription |
1. Initiation 2. Elongation 3. Termination |
|
|
Initiation |
beginning of transcription in initiation, a group of binding proteins called transcripton factors identifies a promoter on the DNA strand |
|
|
Promoter |
sequence of DNA nucleotides that designates a beginning point for transcription "regulatory signal" At the promoter, the transcription factors assemble into a transcription initiation complex, which includes RNA polymerase Promoter sequences help regulate where on the genome transcription can take place, and how often certain sequences are transcribed |
|
|
RNA polymerase |
The major enzyme of transcription |
|
|
Consensus sequence |
The most commonly found promoter nucleotide sequence recognized by a given species of RNA polymerase is called the consensus sequence variation from the consensus sequence causes RNA polymerase to bond less tightly, less often, which leads to associated genes being transcribed less frequently |
|
|
Elongation |
Second step in transcription after binding to the promoter, RNA polymerase unzips the DNA double helix, creating a transcription bubble, after which the complex switches to elongation mode in elongation, RNA polymerase transcribes only one strand of DNA nucleotide sequence into a complimentary RNA sequence -template strand or (-) antisense strand [the other strand (+) sense strand or coding strand protects its partner against degradation] RNA polymerase moves 3' to 5' building in 5' to 3' |
|
|
Errors in transcription |
There is no proof reading mechanism in transcription (not called mutations if they exist) Errors in RNA are not transmitted to progeny; generally not harmful |
|
|
Termination |
last step in transcription end step; occurs when a specific sequence of nucleotides known as the termination sequence is reached Can also involve rho proteins that help to dissociate RNA polymerase from the DNA template |
|
|
Activators and repressors |
activators and repressors bind to DNA close to the promoter and either activate or repress the activity of RNA polymerase often allosterically regulated by small molecules such as cAMP |
|
|
Primary function of genetic control in prokaryotes and eukaryotes |
Prokaryotes- respond to changes in internal and external environment Eukaryotes- homeostasis; control of intracellular and extracellular environments----hallmark of multicellular organisms |
|
|
Jacob-Monod model (watch Khan video) note- prokaryotes |
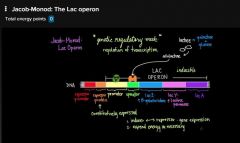
gene regulatory mechanism via transcription Key take-aways: 1. inducer-repressor gene expression 2. Cells only expending energy when necessary
|
|
|
Operon (genetics) |
the genetic unit consisting of the operator, promotor, and genes that contribute to a single prokaryotic mRNA |
|
|
lac operon |
commonly used, and well-studied, example of an operon in E. Coli species Lac operon codes for enzymes that allow cell to import and metabolize lactose when glucose is not readily available Lac operon activated if two conditions are met: 1. glucose is scarce 2. lactose is present |
|
|
Positive control (lac operon) |
in a form of positive control, CAP activates (catabolite activator protein) the promoter, allowing the formation of the initiation complex and the subsequent transcription and translation of three proteins |
|
|
Gene Repression (lac operon) |
when lactose is not present in the cell, a lac repressor protein binds to the operator site and prevents transcription of lac genes, thereby repressing gene expression
|
|
|
Primary Transcript |
Part of post transcription processing by Eukaryotic cells The initial mRNA sequence is not yet ready to be translated until it has been modified by by the cell Primary transcript (pre-mRNA; hnRNA) initial RNA nucleotide sequence arrived at through transcription |
|
|
Purpose of primary transcript |
modifications change the primary transcript (pre-mRNA ; hnRNA) into final processed mRNA helping initiate translation (recognize mRNA) protect mRNA from from degredation eliminate extraneous sequences prior to transl. mechanism for variability from single transcript |
|
|
5' CAP |
before mRNA is fully transcribed, its 5' end is capped in a process using GTP 5' CAP serves as attachment site in protein synthesis during translation protects against degradation by enzymes that cleave nucleotides (exonucleases) |
|
|
Poly A tail |
prior to mRNA being fully transcribed, the 3' end is also protected against exonucleases via addition of a long series of adenine nucleotides when tail is added, 3' end is said to be polyadenylated |
|
|
Splicing |
The primary transcript is much longer than the mRNA that will be translated. before leaving the nucleus, portions of the primary transcript are excised and discarded through a process called splicing Splicing adds exons together to form one continuous sequence of mRNA |
|
|
Introns |
portions of primary transcript that are removed through the splicing process (see splicing) generally much longer than exons |
|
|
Exons |
portions of the primary transcript that become the mature mRNA (see splicing/ introns) |
|
|
snRNPs |
"snurps" The mechanism of splicing involves several small ribonucleoproteins (snRNP's) Each snRNP contains an assortment of proteins ans snRNA snRNP's act as rhibozyme during splicing process (see rhibozyme) |
|
|
Rhibozyme |
and RNA molecule capable of catalyzing specific chemical reactions (one of very few enzymes that is not a protein) |
|
|
Spliceosome |
Splicing occurs when snRNPs recognize nucleotide sequences at the end of introns. snRNPs pull the ends of the introns together, forming a loop or lariat. The complex formed from the association of the snRNPs and the additional associated proteins is called a spliceosome spliceosome excises the introns and joins the ends of the exons together to form mature mRNA that ultimately codes for polypeptide |
|
|
Alternative splicing |
Eukaryotic cells exert control and add variety via alternative splicing- one of the major mechanisms of eukaryotic gene expression regulation allows cell to incorporate different coding sequences into mature mRNA creates a variety of mRNA molecules for translation from a single DNA coding sequence 25,000 protein-coding regions in body, but regulation allows for more than 100,000 proteins |
|
|
Translation |
process by which a cell creates the protein products that are necessary to carry out the business of life translation of nucelotide sequences of mRNA into the amino acid sequence of the corresponding protein |
|
|
Genetic code (translation) |
mRNA nucleotides are strung together to form a genetic code, consisting of four different nucleotides the four RNA nucleotides (adenine, guanine, cytosine, and uracil) together must create a language that unambiguously codes for the 20 common amino acids that make up functional proteins |
|
|
Triplet code |
genetic code of RNA accomplishes it's goal (coding for 20 amino acids) by using a triplet code. series of three nucleotides code for each amino acid series of three only codes for one amino acid, so unambiguous |
|
|
degenerative |
4^1 = 4 amino acids
4^2= 16 amino acids 4^3= 64 amino acids (hence, triplet code) 64 possible, but really only 20, so that means that more than one sequence codes for same amino acid, hence, degenerative |
|
|
Genetic Code - Stop and Start sequences |
Start codon = AUG Stop codon = UAA, UAG, UGA |
|
|
Translation: three major RNA types |
mRNA is the template which carries the genetic code from the nucleus to the cytosol in the form of codons tRNA renders mRNA into a specific amino acid sequence rRNA makes up small and large portion of ribosome (+other associated proteins) |
|
|
Anticodon |
each tRNA has two distinct ends, one end is a series of three nucleotides called the anticodon, which will bind to the complementary codon sequence on the mRNA |
|
|
tRNA |
tRNA renders mRNA into a specific amino acid sequence: two distinct ends - anticodon end / and other end that carries the amino acid that corresponds to the complementary codon sequence on mRNA
non-anticodon end can be added to growing polypeptide chain as tRNAs bind to the codons along the mRNA strand |
|
|
mRNA |
mRNA is the template which carries the genetic code from the nucleus to the cytosol in the form of codons
|
|
|
wobble pairing |
first two base pairs in a codon and anticodon must be strictly complementary (A with U, C with G) Some flexibility in the third base pair, which is called wobble pairing Explains why multiple codons can code for same amino acid Ex: lysine AAA, AAG |
|
|
Ribosome |
all translation takes place via specialized organelle called ribosome free-floating (cytosol) or attached to outer surface of ER composed of small subunit and large subunit made from rRNA and many separate proteins prokaryotic smaller than eukaryotic |
|
|
Svedberg units (S) |
ribosome and subunits are measured in S, which measures sedimentation coefficients via centrifuge, proportional to mass and related to shape and density prokaryotes= 30S + 50S subunits Combined sedimentation coefficient = 70S Eukaryotes= 40S + 60S subunits combined sedimentation coefficent = 80S |
|
|
nucleolus |
complex structure of ribosomes requires a special organelle called the nucleolus to manufacture them prokaryotes do not have a nucleolus, but manufacture in similar manner ribosomes manufactured in nucleolus, however small and large transported into cytosol separately |
|
|
Stages of translation |
similar to transcription 1. initiation 2. elongation 3. termination different in terms of product transcription product = strand of RNA translation product = chain of amino acids |
|
|
initiation factors |
co-factor proteins after mRNA leaves the nucleus via pores, initiation factors help attach the 5' end of mRNA to the small subunit of the ribosome |
|
|
initiation complex |
after mRNA leaves the nucleus, attaches its 5' end to the small subunit of ribosome via co-factors... a tRNA containing 5'-CAU-3' anticodon sequesters AA methionine and settles into the p site (peptidyl site); this is the signal for the large subunit to join and form the initiation complex |
|
|
Initiation (stage) |
the initiation stage is comprised of the initiation factors and the subsequent formation of the initiation complex most of the regulation of translation occurs via the initiation stage through the recognition (or lack of) between ribosomal subunits and secondary structure of mRNA transcript |
|
|
Elongation |
Once the initiation complex is completely formed, we move into the elongation stage during elongation the robosome slides down mRNA, one codon at a time, in the 5' --> 3' direction, matching each mRNA codon to a complimentary tRNA anticodon tRNA, with the input of energy, is able to transfer the language of nucleic acids to the language of amino acids, forming the protein EPA site transitions build the amino acid chain |
|
|
A site |
within each ribosome there are three sites where tRNA molecules can bind. At the A site the Anticodon matches up with the codon, ensuring the right Amino acid is chosen |
|
|
E site |
within each ribosome there are three sites where tRNA molecules bind.
at the E site, the tRNA, which now lacks an amino acid, is free to exit the ribosome |
|
|
P site |
within each ribosome there are three sites where tRNA molecules bind
at the P site, a peptide bond between amino acids is formed, lengthening the polypeptide chain
|
|
|
Termination
|
3rd stage of translation translation ends when a stop codon (nonsense codon) reaches the A site, proteins called release factors (co-factor proteins) bind to the A site, allowing a water molecule to add to the end of the polypeptide chain polypeptide is freed; ribosome breaks up |
|
|
Post-translational modifications |
once the protein is completely synthesized, it can undergo post-translation modifications (sometimes happens in the ER lumen) regulation for expression affect which proteins ultimately become functional addition of sugar, lipid, or phosphate groups to amino acids to influence functionality polypeptide may be cleaved or separate polypeptides may join to form Quaternary structure |
|
|
Signal peptide |
used for determining post-translation ER bound or not ER bound a 20 amino acid sequence, called a signal peptide, near the front of the polypeptide, is recognized by a protein-RNA signal-recognition particle (SRP) |
|
|
Singal-recognition particle (SRP) |
used for determining post-translation ER bound or not ER bound SRP carries the entire ribosome complex to a receptor protein on the ER. There the protein grows across the membrane, where it is either released into the lumen or remains partially attached to the ER SRPs may also target mitochondria, organelles, or other structures; thus, the protein ends up where it needs to be |

