![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
94 Cards in this Set
- Front
- Back
- 3rd side (hint)
|
Membrane |
membrane is 50% lipid and 50% protein - lipids are structure of membrane - proteins carry out membrane function
|
|
|
|
Protein functions in membrane |
transporters: transport charged/large polar molecules across lipid bilayer linkers: membrane protein link proteins on cytosolic face to noncytosolic face anchors: membrane proteins anchor proteins on either side to membrane receptors: detect chemical signals on one face & relay them to other face enzymes: catalyze specific reactions on either side of membrane |
|
|
|
2 types of membrane-associated proteins |
integral: directly attached to membrane peripheral: proteins bound to membrane indirectly only by interactions w/ other membrane proteins |
|
|
|
transmembrane proteins |
integral proteins, which span through bilayer with portions exposed on both sides of membrane transmembrane proteins w/ even # of membrane spanning region has 2 orientation (in-in or out-out) transmembrane proteins w/ odd # of membrane spanning regions has 1 orientation (in-out) transmembrane proteins have hydrophobic & hydrophilic regions -hydrophobic regions are AAs w/ uncharged/non polar side chains & are interior of bilayer -hydrophilic regions are AAs w/ charged/polar side chains & exposed to aqueous env of membrane
|
|
|
|
detergents |
detergents are reagents that solubilize transmembrane proteins & disrupt hydrophobic interactions - are small, amphipathic, lipid-like that contain both hydrophobic (tail) & hydrophilic (head) groups - 2 examples: SDS (strong ionic detergent has ionized grp (sulphate) on head) triton X-100 (mild non-ionic detergent has non-ionized but polar grp at hydrophilic head) - have one hydrophobic tail! - shaped as cones & h2o become clusters called micelles (circle) |
|
|
|
Solubilization of transmembrane proteins by detergents
|
Hydrophobic tails of detergents bind to hydrophobic regions of transmembrane protein & to hydrophobic regions of lipid molecules (separates protein from lipids)
Hydrophilic side of detergents brings binding of membrane proteins into solution as "protein-detergent complexes" |
|
|
|
Proteins
|
AAs are linked by peptide bonds which makes a protein
Peptide bonds that make polypeptide chain are polar & hydrophilic Atoms that form peptide bonds are connected by H bonds with each other |
|
|
|
α-helix proteins
|
1st folding pattern of transmembrane proteins
- Is generated when single polypeptide chain turns around itself to make rigid cylinder! - are receptors for extracellular signals — extracellular part of protein binds signal molecule (ligand) while the cytosolic part signals to cell's interior 1st category α-helices: -AAs are hydrophobic & exposed on outside of helix which contacts with hydrophobic tails of lipid molecules 2nd α-helices: -hydrophobic AAs lie on one side of helix (exposed to lipids) & hydrophilic AAs on other -hydrophilic AA side chains form hydrophilic pore created by packing many helices side by side in "ring" within hydrophobic env of bilayer |
|
|
|
Transmembrane hydrophilic pore
|
5 transmembrane α-helices form h2o-filled pore across bilayer
— selective transport of large polar molecules (AAs, NTs, sugars) & small charged molecules (ions) across membrane |
|
|
|
β-sheet
|
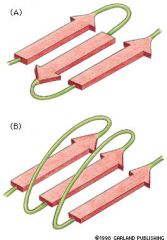
2nd folding pattern of membrane
— polypeptide chains are held together by H bonds between peptide bonds in different polypep chains A) parallel β sheets B) anti parallel β sheets β sheets curve to cylinder which form open-ended "β barrel" – hydrophobic AAs contact hydrophobic core of bilayer – hydrophilic AAs face inside of barrel & "line aqueous channel" Water-filled pores inside β barrel allow selective transport of polar molecules & small charged molecules |
|
|
|
Other integral membrane proteins
|
Are in bilayer but are anchored in bilayer by covalently attached lipids or glycolipids
|
|
|
|
Peripheral membrane proteins
|
– not inserted into hydrophobic interior of bilayer
– are indirectly associated w/ membranes by interactions w/ integral membrane proteins (interactions are by electrostatic bonds) – these proteins dissociate from membrane by being treated w/ polar reagents (high pH/salt concentration) which do not disrupt bilayer! |
|
|
|
Permeability of synthetic lipid bilayer
|
Small hydrophobic molecules (O2, N2, CO2, benzene) YES passes through
Small uncharged polar molecules (H2O, ethanol, glycerol) YES passes Large uncharged polar molecules (AAs, glucose, NTs) NO don't pass Charged molecules (ions) NO don't pass — LUP & charged molecules are water soluble ***transmembrane are permeable to large uncharged polar molecules & charged molecules*** |
|
|
|
Synthetic lipid layer
|
Protein-free artificial lipid bilayers
|
|
|
|
Each transport protein in membrane transfers particular type of molecule.
Organelles also have transport proteins in membrane so? |
Determines exactly what water soluble molecule can pass in/out cell
— organelles have its own characteristic set of transfer proteins |
|
|
|
Concentration of positively charged ions (cations) & negatively charged ions (anions) in cell
|
[inside cations]=[outside cations]=150 mM
[inside anions]=[outside anions]=160 mM To prevent a cell to be torn apart by electrical forces: 1) quantity of pos charge inside cell must be balanced by equal quantity of neg charge 2) quantity of pos charge outside cell must be balanced by equal quantity of neg charge |
|
|
|
Passive diffusion
|
Gases, hydrophobic, small uncharged molecules able to pass membrane
— molecule dissolves in bilayer, diffuse across it & then dissolves in aqueous solution at other side of membrane — no membrane proteins involved & no external source of energy needed Net flow of molecules is "down concentration gradient" (going from more concentration to less) |
|
|
|
Facilitated diffusion
|
— passive transport
Large polar uncharged molecules & ions passes across membrane without directly interacting w/ hydrophobic interior Membrane transport proteins are involved No external energy Net flow of molecule is down concentration gradient |
|
|
|
Active transport
|
Membrane transport proteins involved & external energy needed
Net flow of molecules against concentration gradient Energy needed is provided by another coupled reaction w/ it to drive molecules uphill (like ATP hydrolysis) |
|
|
|
ATP-powered pumps
|
Transport protein
ATPase uses energy of ATP hydrolysis to move H2O-soluble molecule across membrane, against concentration gradient Energy comes from hydrolysis of ATP to ADP & Pi |
|
|
|
Channel proteins
|
Transports specific types of ions down concentration gradient
Forms hydrophilic pore across bilayer through which specific inorganic ions can diffuse Exist in open/close conformation & multiple ions move through it simultaneously when open |
Ion channels
Transport protein |
|
|
Carrier proteins
|
Binds water soluble molecule on one side of membrane & delivers it to other side through change in confirmation of carrier protein
Can only bind few substrate molecules at a time |
Transport proteins
Transporters |
|
|
Carrier proteins
|
Binds water soluble molecule on one side of membrane & delivers it to other side through change in confirmation of carrier protein
Can only bind few substrate molecules at a time |
Transport proteins
Transporters |
|
|
Uniporters
|
Type of carrier protein
Transport single type of molecule down concentration gradient (AAs or glucose across membrane) |
|
|
|
Symporters
|
Type of carrier protein
Couples movement of one type of ion against concentration gradient to movement of different ion down its concentration gradient Moves both ions in same direction across membrane Like ATP-powered pumps, they mediate couple reactions in which energetically unfavourable reaction is coupled/driven by an energetically favourable rxn |
|
|
|
Antiporters
|
Type of carrier protein
Like symporters but instead both ions move in opposite directions across membrane |
|
|
|
Cotransporters
|
Symporters & Antiporters are coupled transporters or "Cotransporters"
|
|
|
|
Na + / K + ATPase
|
Maintains intercellular Na/K concentration in cells
Responsible for coupled rxn of K/Na into or out of cell Hydrolysis of 1 molecule of ATP to ADP & Pi is coupled to export 3 Na ions & import 2 K ions against their concentration gradients across PM |
|
|
|
Mechanism of Na + / K + ATPase in PM
|
Step 1: binding of 3 Na ions
– in E1 conformation, Na/K ATPase has 3 high-affinity Na binding sites & 2 low-affinity K binding sites on cytosolic facing surface of protein – due to high affinity, 3 Na ions bind despite low intracellular concentration – despite high intracellular K concentration, K unable to bind to low affinity site Step 2: binding of ATP – ATP bonds to site on cytosolic side – ATP then hydrolysis to ADP & liberated phosphate is transferred to specific aspartate residue in Na/K ATPase which forms "high energy acyl phosphate bond" (E1-P) Step 3: E1-P conformational change to E2-P & outward transport of 3 Na ions – protein changes conformation to E2-P during the transition 3 bounded Na ions move outward (though protein) – E2-P conformation gives 2 high-affinity K sites on extracellular face Step 4: dissociation of 3 Na & binding of 2 K ions – 3 Na dissociates into extracellular medium from low affinity Na site – 2 K ions bind to K binding site (high affinity despite low concentration) Step 5: hydrolysis of aspartyl phosphate, E2-P conformation change to E1 – aspartyl-phosphate bond in E2-P hydrolyzes, so E2-P is E1 now – during this transition, 2 bounded K ions move inward (cytosol) Step 6: dissociation of 2 K ions – 2 K ions dissociate into cytosol from low affinity sites despite high K concentration |
|
|
|
Properties of ion channels
|
Ion selectivity:
– ion channels are highly selective (narrow pores in channel restrict passage) – pore allows passage of Na bound to single h2o molecule but not K Gating: Ions channels not open permanently, gates open in response to specific stimuli |
|
|
|
Types of ion channels
|
Voltage-gated
—opens in response to change in electrical potential across PM Ligand-gated —open when chemical ligand binds to channel (outside/inside) Stress-activated —open when mechanical force is applied to channel |
|
|
|
CFTR Cl- channel
|
Unusual that it requires both ATP hydrolysis & phosphorylation in order to open Cl- channel in epithelial cell
Recessive mutations in CFTR gene cause cystic fibrosis Cystic fibrosis: Production of thick sticky mucus by several types of epithelial cells, including cells that line respiratory/gastrointestinal tract (excessive salt in sweat) |
|
|
|
For cell to function, where should proteins go?
|
Proteins must go to correct membrane-bound organelle in cell!
Na/K ATPase goes to PM RNA polymerase to nucleus Protease to lysosomes Catalase to peroxisomes ATP synthase to mitochondria Hormones to extracellular space |
|
|
|
Proteins synthesis
|
Few:
– are encoded by DNA in mitochondria/chloroplast – synthesized on ribosomes inside mito/chloro – incorporated to departments in mito/chloro Most: – encoded by nuclear DNA – synthesized on ribosomes in cytosol – delivered to organelle of destination from cytosol |
|
|
|
Protein sorting
|
Process of directing each newly made protein to particular membrane-bounded organelle
Steps in protein sorting: 1) recognition of signal sequence by shuttling cytosolic receptor 2) targeting to outer surface of organelle 3) insertion of targeted protein into membrane/transport across membrane |
|
|
|
Sorting signal
|
Continuous stretch of AAs that directs a protein to (signal sequence) organelle in which it is required
— proteins destined for ER have amino terminal sequence that directs them to organelle needed — protein destined to remain in cytosol lack sequence — if ER signal sequence attaches to cytosol protein, the altered proteins end up in abnormal location in cell |
|
|
|
How does protein cross membranes that are normally impermeable to hydrophilic molecules?
|
Protein is hydrophilic, contains AA which are polar & charged
|
|
|
|
Mechanisms by which membrane-bounded organelle import proteins
|
1) transport through nuclear pores (proteins to nucleus)
– proteins transport through pores that act as selective gates (actively transports only specific proteins) – proteins remain folded during transport 2) transport across membranes (proteins to mitochondria, ER..) – proteins transport across membrane by protein translocators which located in membrane – protein must unfold to get through membrane 3) transport by vesicles (Golgi) – proteins sorted from ER to another compartment of endomembrane system – transport vesicles are loaded w/ cargo of proteins from interior space (lumen) of one compartment (pinch off from its membrane) – proteins remain folded during transportation |
|
|
|
Nuclear pore complexes
|
Made of multiple copies of different proteins
Selectively transports macromolecules across nuclear envelope like large polar molecules, ions, globular proteins (do not diffuse large proteins) NPC nuclear ring supports 8 basket filaments whose distal ends are joined by terminal ring forming structure called nuclear basket Cytosolic ring supports 8 cytosolic filaments Large molecules are actively transported through central plug of NPC (cannot diffuse through H2O filled channels in NPC like ions..) Large proteins need nuclear localization signal (NLS) to pass through central plug |
|
|
|
Mechanism of active protein transport through NPC
|
In cytosol, nuclear import receptors (importins) bind to NLS of cargo protein being transported (binding is driven by energy from GTP hydrolysis)
Importins direct cargo protein to NPC by interacting w/ cytosolic filaments of NPC Binding of cargo protein to NPC opens pore, cargo protein & importins are transformed to nucleus (energy from ATP hydrolysis) Importins-cargo protein complex dissociates (importins are exported back through NPC for reuse) |
|
|
|
Protein import into mitochondrial matrix
|
Cytosolic chaperones (MSF & Hsp 70) use energy of ATP hydrolysis to keep newly synthesized cytosolic proteins in unfolded state, which are delivered to channel-linked receptors in outer mitochondrial membrane
After chaperones released, protein passes through channel in outer membrane & another channel in inner membrane (this translocation into matrix occurs only at contact sites, where outer/inner membrane are in close proximity) Newly imported protein binds to matrix Hsp 70, which uses energy of ATP hydrolysis to allow import without protein folding Hsp 70 is released & matrix targeting sequence is removed by matrix protease Some proteins fold to mature conformation w/ or without help of chaperone (Hsp 60 helps in folding by ATP hydrolysis energy) |
|
|
|
Types of ribosomes in cytosol
|
Membrane-bound ribosomes:
Attached to cytosolic surface of ER membrane & synthesizes proteins that go to ER Free ribosomes: Synthesize a proteins & unattached to membrane |
|
|
|
Translocation of soluble protein across ER membrane into lumen
|
Cytosolic protein, signal recognition particle (SRP) binds to exposed ER signal sequence & ribosome which slows protein synthesis in ribosome
SRP-ribosome complex then binds to SRP receptor in ER membrane SRP & receptor dissociate from ribosome-bound polypeptide chain: - ER signal sequence binds to translocon (protein translocation channel in ER membrane) - ER signal sequence & adjacent segment of growing polypeptide chain, insert as loop into central cavity of translocon ER signal sequence cleaved by signal peptidase in ER lumen ER chaperone (Hsp 70) uses ATP hydrolysis to pull inside into lumen & assists in final folding of protein into mature state |
|
|
|
Transport by vesicles
|
Outward pathway of vesicular transport (secretory pathway)
Proteins transported from ER to golgi to lysosome to membrane/external medium |
|
|
|
Two types of secretory pathways or branches
|
Lysosomal branch
Exocytic branch (exocytosis) |
|
|
|
To function properly, transport vesicles that buds off from compartments must?
|
Take w/ it only proteins appropriate to its destination
Fuse only w/ appropriate target membrane |
Each organelle of endomembrane system maintains its own distinct identity w/ its own distinctive protein composition
|
|
|
Synthesized proteins destined to reside in organelles of endomembrane system are cotransitionally imported to?
|
Rough ER as unfolded, monomeric, unmodified polypeptides
The folding, assembly into multimeric complexes & covalent modification occurs in rough ER |
|
|
|
Synthesized proteins in membrane/lumen of ER undergo 5 principal modifications before reaching final destinations
|
Formation of disulfide bonds
- only formed in lumen of rough ER (oxidative env) not in cytosol (reducing env) - only found in secretory proteins & extracellular domains of PM proteins - function: helps stabilize structure of proteins that encounter changes in pH & degradative enzymes after being incorporated in PM Addition/processing of carbohydrates - proteins that initially enter ER lumen/membrane contain carbohydrate chains - covalent attachment of short carbohydrate chains converts protein to glycoproteins - glycosylation carried out by glycosylating enzymes found in ER only - N-linked protein glycosylation in ER: polypeptide chain enters ER lumen, it's glycosylated by covalent attachment of preformed branched oligosaccharide to particular asparagines in poly - functions of oligosaccharide attached covalently proteins in ER: —protects protein from degradation —retains protein in ER until properly folded —help guide protein to appropriate organelle by serving as transport signal for packaging protein into appropriate transport vesicle Specific proteolytic cleavage Proper folding - correct folding of proteins facilitated by several ER chaperones (proteins that accelerates folding within ER lumen) Assembly into multimeric proteins - folding/assembly of proteins into multimeric protein complexes is assists by ER chaperones (calnexin & calreticulin) |
|
|
|
Steps of folding/assembly of proteins into multimeric protein complexes
|
1) protein glycosylation
2) binding to calnexin/calreticulin & formation 3 disulfide bonds 3) release of ribosome & assembly of trimeric protein |
|
|
|
Mechanisms that controls protein exit from ER
|
1) ER-resident proteins retained in ER by ER retention signal
– signal is carboxyl-terminal tetrapeptide KDEL – KDEL receptor binds to ER retention signal then retains ER-resident proteins in ER after it retrieves protein w/ KDEL sequence that've escaped to cis-Golgi network & return them to ER **proteins that are able to exit: don't contain ER retention signal are properly folded & assembled into multi-subunit protein complexes 2) Quality control in ER – misfolded/unassembled non-resident proteins are actively retained in ER lumen by binding to chaperones – interactions w/ chaperones holds misfolded proteins in ER until proper folding/assembly occurs – proteins that fail to fold properly: are exported from ER to cytosol which are degraded in cytosol **properly folded monomeric/multimeric non-resident proteins are able to exit ER 3) assembly of protein coat on cytosolic surface of transport vesicle – vesicles that bud from ER & others have protein coat on cytosolic surface & are called coated vesicles – after budding is completed, coat is lost so membrane of vesicle to interact directly w/ membrane to which it fuses to |
|
|
|
Mechanisms that controls protein exit from ER
|
1) ER-resident proteins retained in ER by ER retention signal
– signal is carboxyl-terminal tetrapeptide KDEL – KDEL receptor binds to ER retention signal then retains ER-resident proteins in ER after it retrieves protein w/ KDEL sequence that've escaped to cis-Golgi network & return them to ER **proteins that are able to exit: don't contain ER retention signal are properly folded & assembled into multi-subunit protein complexes 2) Quality control in ER – misfolded/unassembled non-resident proteins are actively retained in ER lumen by binding to chaperones – interactions w/ chaperones holds misfolded proteins in ER until proper folding/assembly occurs – proteins that fail to fold properly: are exported from ER to cytosol which are degraded in cytosol **properly folded monomeric/multimeric non-resident proteins are able to exit ER 3) assembly of protein coat on cytosolic surface of transport vesicle – vesicles that bud from ER & others have protein coat on cytosolic surface & are called coated vesicles – after budding is completed, coat is lost so membrane of vesicle to interact directly w/ membrane to which it fuses to |
|
|
|
2 functions of protein coat on cytosolic surface
|
Shapes donor membrane into bud
Helps capture only cargo proteins into budding vesicles |
|
|
|
Components that participate in budding of coated vesicles
|
Budding is started by recruitment of small GTP-binding protein from cytosol to cytosolic surface of donor membrane (GTP-binding protein regulates rate of vesicle formation)
Coat & adapter proteins are recruited from cytosol (cytosolic surface of budding vesicles) Adapter proteins select which membrane proteins will enter transport vesicles as cargo proteins (contain short signal sequences that are recognized by corresponding adapters & directs them to specific type of transport vesicle) Coat subunit proteins polymerizes around cytosolic face of budding vesicle & allows vesicle to pinch off donor organelle GTP-binding proteins hydrolyze their binding GTP & pinch off vesicle |
|
|
|
Types of coated vesicles that transport proteins organelle to organelle
|
COPI vesicle (Golgi to ER)
COPII vesicle (ER to Golgi) Clathrin (Golgi to lysosome) |
Fusion of 3 transport vesicles w/ target membrane have common features:
Fusion occurs after coats are deploymerized Transport vesicle must specifically recognize correct transport membrane Vesicle & target membrane must fuse, to deliver contents between each other |
|
|
Proteins that are involved in fusion of transport vesicles w/ target membrane
|

|
|
|
|
Targeting & fusion of transport vesicles w/ target molecules
|
Docking:
Attachment of transport vesicle to target membrane is mediated by binding v-SNAREs on vesicle/target membrane Fusion: Fusion of 2 membranes requires GTP hydrolysis by Rab proteins Disassembly of SNARE complexes: NSF/SNAP complex uses energy of ATP hydrolysis by NSF to disassembly v/t-SNARE complex (which allow SNARES to be reutilized for another rounds |
Rab GTP-binding proteins needed to form v-SNARE/t-SNARE complexes)
|
|
|
Cell communication
|
Communication occurs by means of extracellular signal molecules
- signal molecules synthesized/released by signalling cells & produce specific response only in target cells w/ receptors for it |
|
|
|
Signal transduction
|
Process of converting an extracellular signal into intracellular signal in target cell
- intracellular signal creates series of rxns that regulates metabolism, movement, proliferation, survival, differentiation of target cell |
|
|
|
Six steps of cell signalling
|
1) Synthesis of signal molecule by signalling cell
2) Release of signal molecule by signalling cell 3) Transport of signal molecule to target cell 4) detection of signal by specific receptor protein 5) change in cellular metabolism, function, development triggered by receptor-signalling molecule complex 6) removal of signalling molecule, terminates cellular response |
Depends on distance, speed & selectivity
|
|
|
Six steps of cell signalling
|
1) Synthesis of signal molecule by signalling cell
2) Release of signal molecule by signalling cell 3) Transport of signal molecule to target cell 4) detection of signal by specific receptor protein 5) change in cellular metabolism, function, development triggered by receptor-signalling molecule complex 6) removal of signalling molecule, terminates cellular response |
Depends on distance, speed & selectivity
|
|
|
Five types of cell signalling
|
Endocrine
Paracrine Autocrine Neuronal Direct cell-cell |
|
|
|
Endocrine signaling
|

|
|
|
|
Endocrine signaling
|

|
|
|
|
Paracrine signalling
|

|
|
|
|
Autocrine signalling
|

|
|
|
|
Neuronal signalling
|

|
|
|
|
Direct cell-cell signalling
|

Contact dependent
|
|
|
|
Direct cell-cell signalling
|

Contact dependent
|
|
|
|
Two types of extracellular signal molecules
|
Molecules are too large/hydrophilic to cross PM so receptors are on outer surface of PM of target cell (cell-surface receptor)
Molecules are small/hydrophobic to diffuse across PM so receptors are in interior of target cell (cytosol/nucleus) — molecules that bind to cell surface receptors which "transduce" (convert) extracellular signals into intracellular signals (which create SIGNAL CASCADES) Signal cascades amplify, distribute & diverge signals received which are distributed to intracellular targets |
|
|
|
Two types of intracellular signal molecules
|
Low molecular weight molecules like cAMP, CA2+ (which are second messenger)
Proteins |
|
|
|
Intracellular signalling cascades have many signal transduction steps
|
Primary signal transduction step:
Receptor protein located on cell surface receives extracellular signalling molecule "ligand" & generates new intracellular signal molecule in response Secondary (downstream) signal transduction step: Intracellular signalling molecules initiate signal cascades |
|
|
|
Classes of ligand-triggered cell surface receptors
|
G-protein-linked
Ion-channel-linked Enzyme-linked |
|
|
|
G-protein linked receptor
|

5) binding to activated G protein activates effector enzyme that generates a specific 2nd messenger
|
|
|
|
Ion-channel-linked recepto
|

|
|
|
|
G-protein linked receptor
|

5) binding to activated G protein activates effector enzyme that generates a specific 2nd messenger
|
|
|
|
Ion-channel-linked recepto
|

|
|
|
|
Enzyme-linked receptor
|

|
|
|
|
G-protein linked receptor
|

5) binding to activated G protein activates effector enzyme that generates a specific 2nd messenger
|
|
|
|
Ion-channel-linked receptor
|

|
|
|
|
Enzyme-linked receptor
|

|
|
|
|
Molecular switches
|
Cascades are formed by intracellular signal proteins that act as molecular switch
3 types Protein phosphorylation-dephosphorylaton switches GTP binding hydrolysis switch Assembly-dissembly of multi protein complexes |
|
|
|
Protein phosphorylation-dephosphorylation switches
|

|
|
|
|
GTP binding-hydrolysis switch
|

|
|
|
|
GTP binding-hydrolysis switch
|

|
|
|
|
Assembly-disassembly of multi protein complexes
|

|
|
|
|
GTP binding-hydrolysis switch
|

|
|
|
|
Assembly-disassembly of multi protein complexes
|

|
|
|
|
Receptors interior of target cell
|
In cytosol
–Complex of receptor/signalling molecule are transported from cytosol to nucleus –complex of receptor/signalling remains in cytosol Extracellular signal molecules that take part of this process are steroid/thyroid hormones Intracellular receptors are: -Proteins known as steroid receptor superfamily -transcription factors that activate/repress transcription of their target genes |
|
|
|
Steroid hormone receptor
|
Bind DNA as diners which contains 4 domains
1) ligand binding domain 2) DNA binding domain 3) dimerizatiom domain 4) transcription activation domain |
|
|
|
Steroid hormone receptor
|
Bind DNA as diners which contains 4 domains
1) ligand binding domain 2) DNA binding domain 3) dimerizatiom domain 4) transcription activation domain |
|
|
|
Gene regulation by steroid hormone "thyroxine & its receptor)
|

|
|
|
|
Steroid hormone receptor
|
Bind DNA as diners which contains 4 domains
1) ligand binding domain 2) DNA binding domain 3) dimerizatiom domain 4) transcription activation domain |
|
|
|
Gene regulation by steroid hormone "thyroxine & its receptor)
|

|
|
|
|
Gene regulation by steroid hormone "cortisol & its receptor)
|

|
|

