![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
30 Cards in this Set
- Front
- Back
|
BIOCHEMISTRY
|
.
|
|
|
Thermodynamics
|
^(change in) G (free energy) = H (enthalpy-potential energy) - T (temp)^S (entropy)
(-)^G = spontaneous reaction: decrease ^H, increase ^S (+) ^G = Non-spontaneous reaction ^G = 0: equilibrium |
|
|
Reaction Coordinate Graph
|
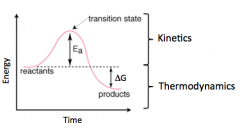
|
|
|
Activation Energy (Ea)
|
Energy required to tech transition state (TS)
Increased Ea = Decreased rate of reaction |
|
|
Catalyst
|
Increases rate of reaction
Stabilizes transition state |
|
|
Key Words
|
Thermodynamics: Spontaneous, ^G, Reactants-->Products
Kinematics: Catalyst, Ea, rate |
|
|
Enzymes
|
- Biological catalyst
- 3 main functions 1) increases rate of reaction 2) does not get used up 3) highly specific - Structure: Proteins (99.8%) |
|
|
Regulations
|
1) Phosphorylation
2) Proteolytic Cleavage 3) Allosteric Regulation 4) Association without polypeptides 5) Feedback Inhibition |
|
|
1) Phosphorylation
|
Covalent Modification
Kinase: Takes P from ATP Phosphorylases: Free floating Pi |
|
|
2) Proteolytic Cleavage
|
Cleaves an enzyme with a protease
Zymogenens: Digestive enzymes |
|
|
3) Allosteric Regulation
|
Allosteric regulator
|
|
|
4) Association without polypeptides
|
Cooperatively: hemoglobin
|
|
|
5) Feedback Inhibition
|
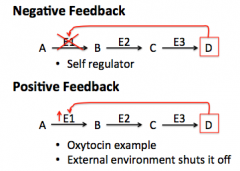
|
|
|
Basic Kinetics
|
Rate of the product forming
Reaction rate (V): rate of product per unit of time |
|
|
Enzyme Kinetics
|
![Enzyme concentration [E] never changes
1) [E] >> [S]
2) [E] = [S]
3) [E] << [S] - saturated: all enzyme active sites filled
Vmax = Saturation reached
Km = Vmax/2 : The affinity the substrate has for the enzyme](https://images.cram.com/images/upload-flashcards/69/46/37/2694637_m.png)
Enzyme concentration [E] never changes
1) [E] >> [S] 2) [E] = [S] 3) [E] << [S] - saturated: all enzyme active sites filled Vmax = Saturation reached Km = Vmax/2 : The affinity the substrate has for the enzyme |
|
|
Cooperativity
|
![Hemoglobin not an enzyme but acts like one!
1) [E] >> [S] - hard to take off
2) [E] = [S]
3) [E] << [S]
As Km increases = affinity decreases
Low affinity hard to bind](https://images.cram.com/images/upload-flashcards/69/47/81/2694781_m.png)
Hemoglobin not an enzyme but acts like one!
1) [E] >> [S] - hard to take off 2) [E] = [S] 3) [E] << [S] As Km increases = affinity decreases Low affinity hard to bind |
|
|
Inhibitors
|
Reduce the amount of product formed
Assume reversibility 2 Types 1) Competitive 2) Non-competitive |
|
|
1) Competitive Inhibitor
|
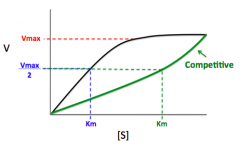
Binds to active site
Vmax stays the same Km will increase |
|
|
2) Non-competitive Inhibitor
|
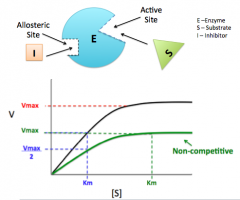
Binds to allosteric site
Vmax changes Km stays the same |
|
|
CELLULAR RESPIRATION
|
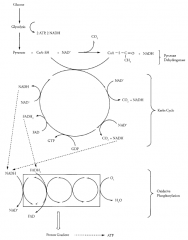
|
|
|
Oxidation
|
1) lose H+
2) lose e- 3) gain O2 |
|
|
Reduction
|
1) gain H+
2) gain e- 3) lose O2 |
|
|
Glycolysis
|

Location: Cytoplasm
O2 Requirement: Anaerobic Molecules formed/used (ATP equivalents): -2 ATP (-2 ATP) 4 ATP (4 ATP) 2 NADH (3 ATP in eukaryotes, 5 ATP in prokaryotes) |
|
|
Pyruvate Dehydroginase Complex (PDC)
|
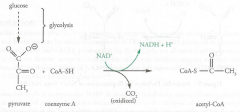
Location: Matrix of mitochondria
O2 Requirement: Aerobic (indirect) Molecules formed/used (ATP equivalents): 2 NADH (5 ATP) |
|
|
Krebs Cycle
|
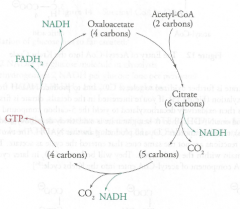
Location: Matrix of mitochondria
O2 Requirement: Aerobic (indirect) Molecules formed/used (ATP equivalents): 6 NADH (15 ATP) 2 FADH2 (3 ATP) 2 GTP (2 ATP) |
|
|
Electron Transport Chain
|
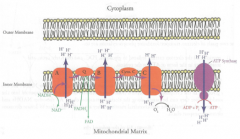
Location: Inter membrane of mitochondria
O2 Requirement: Aerobic (direct) |
|
|
Oxidative Phosphorylation
|
Location: Inter membrane of mitochondria
O2 Requirement: Aerobic (direct) |
|
|
# of protons = 1 ATP
??? |
4 H+ = 1 ATP
|
|
|
Fermentation (no oxygen)
|
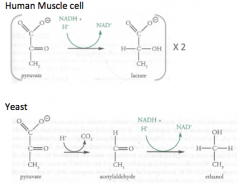
|
|
|
Starvation
|
B (beta) - Oxydation
Takes place in mitochondria Breaks down lipids (fatty acids) 2C chains break down --> goes directly to Krebs cycle |

