![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
98 Cards in this Set
- Front
- Back
|
3 step mechanism |
1. protein synthesis and translocation across ER membrane 2. protein folding and modification in the ER 3. protein transport to the Golgi, lysosome, cell surface, etc |
|
|
rough ER |
1. ER densely studded with ribosomes 2. produces rough microsomes |
|
|
ribosomes |
produce secreted and integral membrane proteins |
|
|
rough microsomes |
1. produced in RER 2. produced when cells are homogenized 3. small, closed vesicles with the same orientation as the RER 4. used to develop assay for protein translocation into ER |
|
|
how are proteins translocated into the ER? |
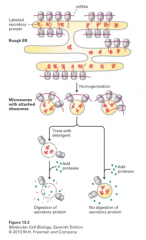
via microsomes |
|
|
secretory proteins |
1. use signal sequences 2. glycolysated in the ER and golgi |
|
|
cell-free protein synthesis |
1. combined with microsomes used in experiments 2. show signal sequences, cotranslational translocation, and maturation, etc. |
|
|
cell-free protein synthesis; no microsomes present |
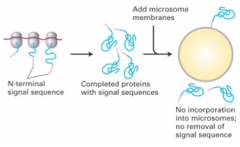
|
|
|
cell-free protein synthesis; microsomes present |
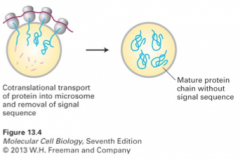
|
|
|
how are signal sequences used? |
1. Two GTPase proteins 2. SRP particle 3. SRP receptors |
|
|
GTPase proteins |
1. critical for using the signal sequences |
|
|
SRP |
1. signal recognition particle 2. cytosolic ribonucleoprotein particle
|
|
|
SRP receptor |
1. docking protein 2. found on ER membrane |
|
|
translocon |
1. Sec61 complex 2. contains a central channel lined with hydrophobic residues 3. allows for transit of an unfolded protein chain while remaining sealed to ions and small hydrophilic molecules |
|
|
60S ribosomal subunit |
1. binds to a protein lined protein channel 2. allows transfer of nascent protein chains via ER membrane |
|
|
Sec61-alpha protein |
1. binding to the nascent peptide 2. identified via crosslinking |
|
|
what drive secretory protein translocation? |
translation |
|
|
component proteins |
1. can be purified and reconstituted 2. those required for co-translational translocation are NOT ATPases |
|
|
what powers translocation through the ER? |
energy of translocation and extension nascent peptide |
|
|
post-translational translocation |
1. proteins can translocate via the ER after full protein synthesis 2. powered by ATP hydrolysis by BiP binding
|
|
|
what powers translocation? |
ATP hydrolysis via BiP binidng |
|
|
Bip binding |
1. chaperone protein binding 2. powers translocation via ATP hydrolysis 3. stabilizes the entering polypeptide |
|
|
types of membrane proteins |
1. Type 1 2. Type 2 3. Type 3 4. Tail-anchored protein 5. Type 4 6. GPI-linked protein |
|
|
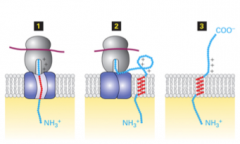
Type 1 membrane proteins |
LDL receptor Insulin receptor GH receptro hydrophobic membrane spanning sequences act as stop transfer signals |
|
|
Type II membrane proteins |
Transferrin receptor Golgi galactosyl-transferase Asialoglycoprotein receptor lack a signal sequence inverted orientation as Type I |
|
|
Type III membrane proteins |
cytochrome p450 lack a signal sequence stop transfer sequence located near amino terminus |
|
|
Tail-anchored membrane proteins |
v-SNAREs t-SNAREs |
|
|
Type IV membrane proteins |
G-protein coupled receptors Glucose transporters Voltage gated Ca channels ABC small molecule pumps Cl- channel Sec61 multipass |
|
|
GPI-linked membrane protein |
plasminogen activator receptor Fasciclin II |
|
|
Type I |
|
|
Type II |
|
|
Type III |
|
|
how are putative membrane spanning sequences identified? |
by scanning sequences in the genome for stretches of 20 hydrophobic amino sequences |
|
|
hydropathy profile |
1. assigning a number for hydrophobic vs charged amino acids 2. allows us to graphically show membrane spanning domains |
|
|
ER |
protein modifications, folding and quality control location of 1. secretory protein glycolysation 2. disulfide bond formation 3. folding and multisubunit complex assembly 4. specific proteolytic cleavages |
|
|
where does secretory protein glycolysation occur? |
ER and Gogli |
|
|
where are disulfide bonds formed? |
ER |
|
|
where does folding and multisubunit complex protein assembly occur? |
ER |
|
|
where do specific proteolytic cleavages occur? |
in secretory pathways ER, Golgi, secretory vesicles |
|
|
protein glycolysation |
1. occurs in ER 2. either O-linked or N-linked
|
|
|
O-linked oligosaccharides |
1. have carbohydrate chains covalently linked to hydroxyl groups 2. serine and threonine residues 3. generally short 4. often contain only 1-4 sugar residues |
|
|
N-linked oligosaccharides |
1. have carbohydrate chains covalently attached to amide groups 2. asparagine residues 3. 14 residue 4. preformed precursor assembled and added in ER; transferred via oligosaccharyl transferase activity 5. contain core with 2 N-acetylglucosamine and at least 3 mannose residues 6. long with several branches |
|
|
N-linked precursor assembly and transfer |
1. biosynthesis of precursor oligosaccharide 2. attached via pyrophosphate linkage to dolichol phosphae 3. transferred to forming proteins via oligosaccharyl transferase activity 4. immediately modified 5. remove all 3 glucose sugars and one specific mannose in the structure |
|
|
dolichol phosphate |
RER membrane lipid that links to pyrophosphate in precursor oligosaccharide biosynthesis |
|
|
what is the consequence of tunicamycin treatment? |
misfolding proteins: Flu hemagglutinin protein |
|
|
disulfide bonds |
1. formed in ER 2. do not for spontaneously in cells under most conditions 3. formed and rearranged via protein disfulide isomerase (PDI) activity 4. added to many soluable secretory proteins and exoplasmic domain of membrane proteins in ER |
|
|
quality control |
chaperones and other ER proteins |
|
|
what are required for proper folding and assembly |
1. BiP, lectins, PDI, and other proteins 2. required for ER exit |
|
|
peptidyl-prolyl isomerase |
1. enzyme catalyzes threotation of exposed peptidyl-propyl bonds 2. can be indiscriminate or substrate specific |
|
|
unfolding protein response |
1. increased unfolded protein levels in ER 2. due to heat shock 3. induced by BiP dissociating from Ire1 and binding unfolded proteins 4. cleavage of Hac1 message 5. produces Hac1 transcription factor 6. induces expression of ER stress protein response |
|
|
Unbound Ire1 |
1. can homodimerize 2. dimer has endonuclease activity |
|
|
nuclear transport |
molecules and large structures are moved in and out of the nucleus via the nuclear pore complex
|
|
|
nuclear pore complex |
1. moves molecules and large structures in and out of the nucleus 2. composed of nucleoporins (FG-repeats) 3. allow passive diffusion of: ions, small metabolites up to 40kD 4. facilitates transport of proteins and complexes like ribosomes across nuclear membrane 5. located in nuclear envelope
|
|
|
nucleoporins |
1. compose nuclear pore complexes 2. assemble into nuclear basket |
|
|
nuclear basket |
1. assembled by nucleoporins 2. associates with the nuclear lamina |
|
|
nuclear lamina |
1. cytoskeletal coat on the inner nuclear membrane 2. associates with the nuclear bakset |
|
|
importins |
1. binds to nuclear protein in the cytoplasm 2. produces a bimolecular cargo complex that is transported through the nucleus 3. driven by GTP hydrolysis |
|
|
NLS |
1. nuclear localization signal 2. directs selective transport to the nucleus
|
|
|
nuclear proteins |
1. synthesized in the cytoplasm 2. contain a NLS 3. bound by importin to produce bimolecular cargo complex |
|
|
bimolecular cargo complex |
1. formed when importin binds to nuclear protein in the cytoplasm 2. translocate into nucleus 3. via stimulation of Ran GTP |
|
|
Ran-GTP/Ran-GDP |
cycles to regulate cargo complex assembly |
|
|
what drives nuclear import? |
GTP hydrolysis |
|
|
Exportins |
1. similar to importins 2. use NES signals and trimolecular (vs bimolecular in importins) during export 3. mRNP exporter 4. does not require Ran-GTPase 5. uses Dbp5, RNA helicase instead 6. requires ATPase activity |
|
|
NES |
1. nuclear export signals 2. stimulate export of proteins that shuttle in and out of nucleus |
|
|
mRNP exporter |
1. used in Nuclear export 2. NXF1/NXT11 dimers 3. function as transport receptors that interact with FG-repeats |
|
|
cytosolic ribsomes |
1. beginning site for: synthesis of secreted proteins, integral plasma-membrane proteins, and proteins destined for the ER, Golgi complex, or lysosome 2. become attached to the ER membrane --> forms RER |
|
|
ER signal sequences on nascent secretory protein |
1. consist of segment of hydrophobic amino acids 2. located at N-terminus |
|
|
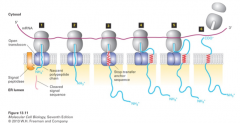
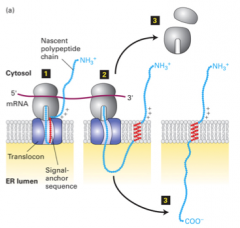
co-translational translocation |
1. SRP recognizes ER signal sequences 2. binds ER signal sequence to nascent secretory protein 3. nascent secretory protein (with bound ER signal) bound to SRP receptor on ER membrane 4. targets ribosome/nascent chain complex to ER 5. SRP and SRP receptor insert nascent secretory protein into translocon 6. SRP and SRP receptor dissociate 7. ribosome attached to translocon continues translation 8. unfolded ER protein chain is extruded into ER lumen 9. no additonal energy required for translocation |
|
|
SRP and SRP receptor |
1. mediate insertion of nascent secretory protein into translocon 2. dissociation: hydrolysis of 2 molecules of GTP by SRP and SRP receptor 3. ribosome attached to translocon |
|
|
ratcheting mechansim |
rquires ATP hydrolysis via BiP chaperone |
|
|
post-translocation mechanism |
1. completed secretory proteins is targeted to ER membrane via interaction of signal sequences with translocon 2. polypeptide chain pulled into ER via ratcheting mechansims
|
|
|
driving force for post-transcriptional translocation in bacteria? |
SecA |
|
|
SecA |
cytosolic ATPase that pushes polypeptides through the translocon channel |
|
|
what happens in both co- and post- translational translocation |
signal peptidase in ER membrane cleaves ER signal sequences from a secretory protein soon after N-terminus enters the lumen |
|
|
integral membrane proteins |
1. synthesized on RER 2. 5 topological classes as well as lipid-linked type |
|
|
topogenic sequences |
1. N-terminal sequences, internal stop-transfer anchor, internal signal-anchor 2. direct the insertion and orientation of nascent proteins within the ER membrane 3. orientation is retained during transport of the completed membrane protein to its final destination |
|
|
single pass proteins |
1. contain one or more topogenic sequences 2. type 1,2, 3 |
|
|
multipass proteins |
1. each alpha helical segment can function as an internal topogenic sequences 2. depending on location in polypeptide chain and presence of adjacent positively charged residues |
|
|
cell-surface proteins |
1. initially synthesized as type I proteins on ER 2. then cleaved with their luminal domain transferred to a GPI anchor |
|
|
Formation of N-linked oligosacchardies |
1. assembly of conserved 14 residue high mannose precursor on dolichol 2. preformed oligosaccharide is transferred to a specific asparagine residue of nascent polypeptide chains in ER lumen 3. 3 glucose residues and 1 mannose residue is removed |
|
|
oligosaccharide side chains |
1. assist in the proper folding of glycoproteins 2. help protect mature proteins from proteolysis 3. participate in cell-cell adhesion 4. function as antigens |
|
|
PDI |
1. protein disulfide isomerase 2. present in ER lumen 3. catalyzes both the formation and rearrangement of disulfide bonds |
|
|
BiP, lectins, PDI, and other proteins
|
1. required for proper folding and assembly 2. assemble in ER |
|
|
what are transported from RER to Golgi? |
only properly folded proteins and assembled subunits |
|
|
what happens to unassembled or misfolded proteins in ER? |
1. transported back to cytosol 2. degraded in the ubiquitin/proteasome pathway |
|
|
FG-nucleoporins |
1. contain FG-repeats 2. line central transporter channel 3. play a role in transport of all macromolecules via nuclear pores |
|
|
how are larger marcomolecules transported through nuclear pores |
via assistance of nuclear transport receptors |
|
|
nuclear transport receptors |
1. transport larger molecules through nuclear pores 2. interact with both the transported molecule and FG-repeats of FG-nucleoporins |
|
|
nuclear restricted proteins |
contain a NLS but not an NES |
|
|
what kind of proteins contain NLS but not NES? |
nuclear restricted proteins |
|
|
what kind of proteins contain both NLS and NES? |
proteins that shuttle between the nucleus and cytoplasm |
|
|
transient interactions between nuclear transport receptors and FG-repeats |
allow very rapid diffusion of nuclear transport protein-cargo complex via the central channel of NPC |
|
|
Ran |
1. G protein that exists in different comformations when bound to GTP or GDP 2. causes unidirectional nature of protein export and import through nuclear pores |
|
|
GEF |
1. localized in nucleus 2. guanine nucleotide-exchange factor 3. Ran GDP --> Ran GTP/cargo complex in nucleoplasm to export in cytoplasm |
|
|
GAP |
1. localized in cytoplasm 2. GTPase activating protein 3. Ran GTP complex --> Ran GDP and cargo in cytoplasm for import
|
|
|
Ran GTP |
1. high in nucloplasm 2. stimulates cargo complexes |
|
|
Ran GDP |
high in cytoplasm |
|
|
draw GTP/GDP mechanism |
on board |

