![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
52 Cards in this Set
- Front
- Back
|
Overview of RNAs |
Messenger: encodes AA sequences of all polypeptides in cell Transfer: match anticodon to mRNA while carrying an AA (for protein synthesis) Ribosomal: constituents of the large and small ribosomal units Micro: regulate expression of genes Ribozymes: catalytic RNA molecules act as enzymes |
|
|
Overview of RNA metabolism |
Transcribed from DNA (tightly regulated transcription) Ribozymes are single stranded, fold into compact structures Processing of mRNA: splicing (introns and exons), poly-adenylation (3’) nucleosiand capping (5’) |
|
|
Transcription in e.coli |
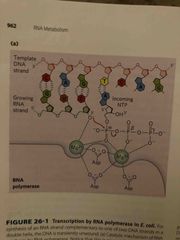
Nucleoside triphosphate add to 3’ of growing RNA strand (it’s complementary to dna template) Synthesis catalyze by RNA polymerase (enzyme) and needs 2 Mg ions to activate RNA polymerase unwinds 17 bp of DNA and covers 35 bp |
|
|
To begin transcription in e.coli |
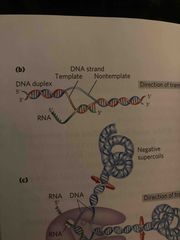
RNA polymerase binds to Promoter to begin (primer not needed) New RNA base pairs with “bubble” in dna |
|
|
RNA polymerase and positive coils |
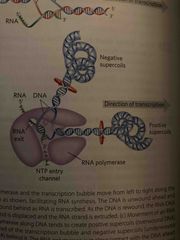
Due to strain at the ends of strands from unwinding, RNA polymerase generates positive supercoils ahead Topoisomerases relieves them later |
|
|
Template vs coding strand |

DNA template strand: serves as template for RNA polymerase DNA coding strand: non-template, has same sequence as RNA transcript |
|
|
Both Dna strands code for proteins |

Coding info may happen on either strand Each strand codes for a number of proteins Adenovirus (common cold) has linear genome |
|
|
RNA polymerase (crab claw) |
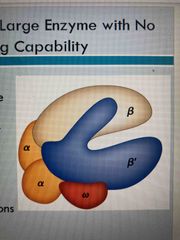
Large holoenzyme 5 subunits: 2 alpha, 2 beta, and psi, plus a sixth subunit Lacks proofreading ability and exonuclease so high error rate Bonds to promotor regions to start transcription |
|
|
The six subunits of RNA polymerase functions |
2 alpha: assembly and binding to upstream promoter 2 beta: 1 is catalytic and other does dna binding The w: protects polymerase from denaturation The sigma: directs enzyme to promoter (each class of RNA polymerases have different sigma subunits) |
|
|
Common features of promoters in e.coli |
TATA sequences: Sigma subunit bonds at -10(TATAAT) and -35(TTGACA) Alpha subunit bonds to upstream promoter btw -40 and -60 These sequences Affect gene expression Nucleotides before the first one of RNA molecule are considered upstream and given negative values
|
|
|
Footprint technique |
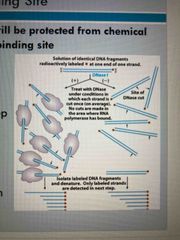
Helps to find dna binding site 1. Isolate dna fragment that might have binding site 2. Radiolabel dna 3. Bind protein to dna and keep other naked as a control group 4. Treat both samples with enzyme to cleave dna 5. Use gel electrophoresis to separate fragments |
|
|
Initiation of transcription (e.coli) |
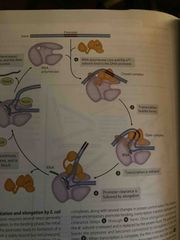
RNA polymerase bonds to promoter with sigma unit (creates a closed complex) An open complex forms RNA polymerase moves away from promoter(clearance) and sigma is replaced by protein NusA |
|
|
Elongation of transcription |
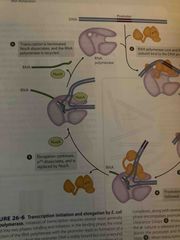
RNA polymerase bonds to triphosphate nucleoside and generates RNA transcript NusG binds to both rna polymerase and ribosome linking them together (affects rate of transcription in prokaryotes) |
|
|
Regulation of transcription |
Transcription is E-intensive so Regulation is achieved in different ways: Decrease affinity of rna polymerase to promoter by… -Activator proteins cAMP receptor protein (CRP) -Repressor proteins block binding sites |
|
|
2 types of termination in e. Coli |
1. p-independent 2. p-dependent |
|
|
p-independent |
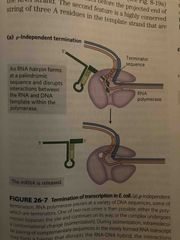
3 U’s at end of transcription (UUU) Self-complementary regions so forms hairpin which causes RNA polymerase to stop and dissociate |
|
|
p-dependent |
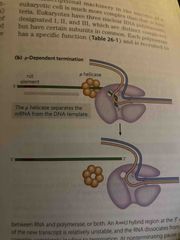
Common CA-rich sequence called a rut site (Rho utilization) p-protein (helicase that binds to rut site) processes until termination site is reached |
|
|
Eukaryotes RNA polymerases |
RNA polymerase I: synthesizes pre-ribosomal RNA (precursor) RNA polymerase II: synthesis of mRNA, very fast, inhibited by mushroom toxin, can recognize 1,000 of promoters RNA polymerase III: makes tRNA and small rna products RNA IV: in plants Mitochondria have their own rna polymerase |
|
|
Eukaryotic mRNA transcription involves many proteins |
Relies on protein-protein contacts RNA polymerase II is a large complex with 12 subunits (some subunits are homologous to bacterial rna polymerase) |
|
|
Assembly of RNA polymerase II at promoter |
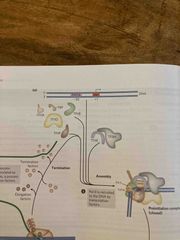
TATA-binding protein (TBP) and promoter initiate assembly Helicase activity in TF2H unwinds dna at promoter Kinase activity in TF2H phosphorylates polymerase carboxyl-terminal domain (CTD), changing conformation and allowing rna polymerase II to transcribe |
|
|
Transcription steps 1-3 |
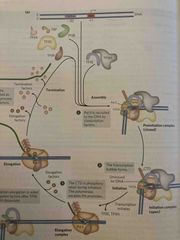
See pic |
|
|
Transcription steps 3-5 |
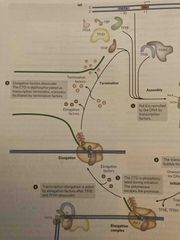
See pic |
|
|
Elongation and termination |
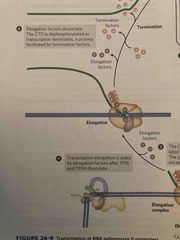
After 60-70 nucleotides, TF2E is released, followed by TF2H Elongation factors bound to rna polymerase II enhance processivity and coordinate posttranslational modifications Polymerase II is de phosphorylated for termination |
|
|
TF2H and repair |
Has role in nucleotide-excision repair (recruits NER complex at lesion) Genetic repair diseases are associated with TF2H Transcribed genes are repaired more than silent genes |
|
|
Inhibition of rna polymerases (not on exam) |
Actinomycin D & acridine: prevents transcription Rifampicin: binds to beta subunit of bacterial RNA polymerase Alpha-amanitan in certain mushrooms blocks polymerase II and III but does not block own II |
|
|
Processing of mRNA overview |
Dozens of proteins coordinate with each other As well as with transport proteins for transporting ribosomes Primary transcript: a new synthesized rna molecule Processing includes: -splicing out introns and joint exons -adding 5’ cap -adding 3’ poly A tail -degradation |
|
|
The 5’ cap |
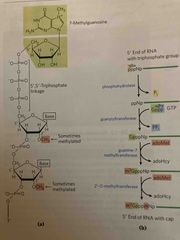
7-methylgaunosine links to 5’ end via 5’,5’ triphosphate link formed using GTP May include additional methylations following cap (methyl groups are from SAMe) Protects RNA nucleases Forms a binding site for ribosome |
|
|
Maturation of mRNA in eukaryotes |
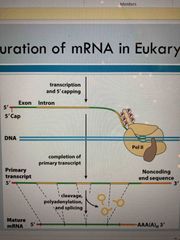
See pic |
|
|
Introns |
Found in most genes Exons are <1,000 bp in length, introns are 50-700,000bp Some genes have dozen of introns Human genome has more than 200,000 introns spread across 20,000 genes |
|
|
Classes of introns |
Groups I and II: self-splicing so no need for proteins or ATP; I’m nuclear, mitochondrial and chloroplast genomes; I’m some bacteria. Group I and II splice differently Spliceosomal introns: spliced by large complexes (spliceosomes); use same mechanism as group 2. These are most common. tRNA introns: spliced by protein-based enzymes |
|
|
Group I introns |
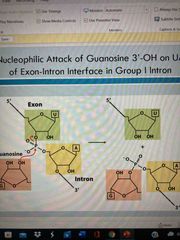
Nucleophilic attack of gaunosine 3’ OH of exon-intron interface Attacks phosohodiester bond btw U and A at end of intron Release U ending exon portion 3’ OH of U-end exon then attacks 5’ of other exon to rejoin pieces |
|
|
Group II introns |
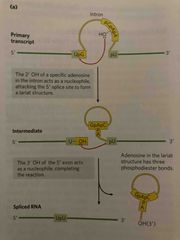
The nucleophile is a 2’ OH of an A residue within the intron After cleavage, 2nd piece forms a horseshoe shape intermediate with a 2’-5’ phisphodiester bond |
|
|
Spliceosome introns |
Removed by spliceosomes Spliceosomes are made of small nuclear ribonuclear proteins (snRNPs or snurps) There are 5 snurps in eukaryotes GU at 5’ end and AU at 3’ end mark site fo splicing |
|
|
Spliceosome introns |
Removed by spliceosomes Spliceosomes are made of small nuclear ribonuclear proteins (snRNPs or snurps) There are 5 snurps in eukaryotes GU at 5’ end and AU at 3’ end mark site fo splicing |
|
|
Addition of poly A tail |
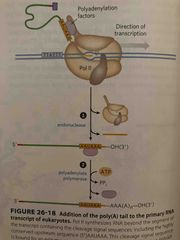
Serves as a binding site on mRNA and protects it from degradation RNA polymerase II synthesizes RNA beyond the cleavage signal sequence Endonuclease cleaves downstream to highly conserved AAUAA Polyadenylate polymerase synthesizes |
|
|
Overview of mRNA processing |
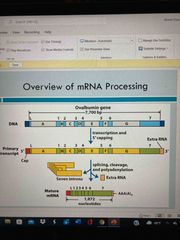
See pic |
|
|
Alternative splicing (not on exam) |
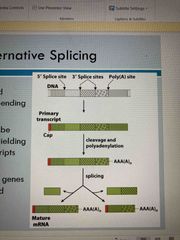
A single gene can yield different peptides Particular regions may be retained or removed yielding different transcripts At least 95% of human genes are alternatively spliced |
|
|
Poly A site choice (not on exam) |
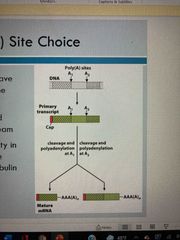
Some transcripts have more than one site where poly A tail forms Site near the 5’ end removed sequence downstream Generates diversity (ie in variable heavy chain of immune-globulin) |
|
|
Alternative splicing in thyroid and brain (not on exam) |
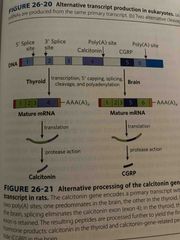
In thyroid will splice so you have calcitonin In brain will splice so you have GCRP All are made from same mRNA |
|
|
Processing of tRNA (not on exam but on MCAT) |
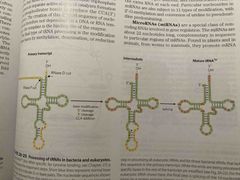
Cap changes from UUA to ACC And splicing reduces bottoms of tRNA See pic |
|
|
MicroRNAs (miRNAs) (not on exam but on MCAT) |
Short noncoding RNAs bind to specific regions of mRNA to alter translation through cleaving or blocking mRNA translation About 1500 human genes encode miRNAs and 1 or more affect the expression of most protein-coding genes Synthesis from larger precursors (drosha & diver) |
|
|
Steps in miRNA processing (not on exam but on MvAT) |
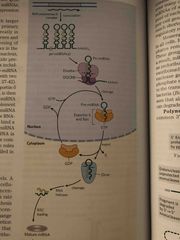
Long precursor called pri-miRNA is made in nucleus and then cleaved by drosha Exportin and Ran protein export to cytoplasm Dicer cleaves into dsRNA Helicase unzips and complement miRNA is loaded onto a protein complex called RNA-induced silencing complex (RISC) |
|
|
RISC miRNA prevents translation of mRNA (not on exam but on MCAT) |
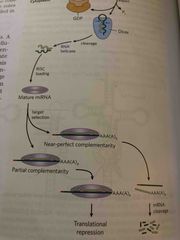
The miRNA sequence in RISC binds to a complement in target mRNA If perfect complement, then target mRNA is cleaved If partial complement, translation is blocked |
|
|
Ribozymes |
RNA molecules that cleave RNA 3-D structure so Inactive if denatured Follows Michaelis-menton rules: Saturable, active site, measurable km, competitive inhibited, nucleophilic attack |
|
|
Examples of ribozymes |
Self-splicing group I introns Hammerhead ribozyme: cleaves viruses (specific to plants) RNaseP: cleaves precursors to tRNA helps to eliminate that extra length of tRNA |
|
|
Degradation of cellular mRNAs |
RNA lifetime is unique to each gene products needs Half-life varied from seconds to hours Degradation occurs via ribonucleases RNA in Bacteria have shorter lives but can be extended if hairpins in structures In eukaryotes, 5’ end and 3’ poly A tail aid in stability of mRNA |
|
|
Reverse transcriptase |
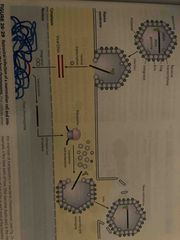
Makes DNA from RNA, then degrades RNA from DNA/RNA hybrid, replacing it with DNA DNA can then be incorporated into the host cell Retroviruses (HIV) Does this in 3 steps: 1.RNA-dependent DNA synthesis 2. RNA degradation 3. DNa-dependent DNA synthesis
|
|
|
Retroviruses contain 3 genes and a long terminal repeat (LTR) |
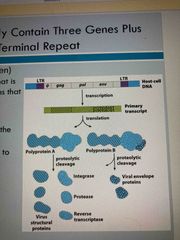
Gag (group associate antigen) Pol (polypeptide): encodes protease, integrase (insert into host) and reverse transcriptase Env: encodes viral envelope LTR allows for integration into host’s DNA |
|
|
Retrotransposons |
In eukaryotes, Similar to retroviruses Encode a protein with Homology to reverse transcriptase Use enzyme to make DNA from RNA |
|
|
Telomeres |
Structures at end of eukaryotic chromosomes Have tandem repeats of T 1-4 and G 1-4 with AC on opposite strand TG strand is longer than complement leaving 3’ end overhang Not easily replicated So chromosomes are shortened with reach generation |
|
|
Telomerase |

Protect ends of chromosomes from shortening Telomerase has RNA with C-A repeat to serve as a template for synthesis of T -G strand of telomere Binds to 3’ end and hangs off to template extends beyond it Gap on bottom strand is filled by DNA polymerases |
|
|
H |
H |

