![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
245 Cards in this Set
- Front
- Back
|
What is a polymer of nucleotides?
|
DNA
|
|
|
What makes up nucleotide?
|
Phosphate + Sugar + Base
|
|
|
Nucleoside
|
Base + Sugar
|
|
|
Purines
|
AG
|
|
|
Pyrimidines
|
TCU
|
|
|
nucleoside/nucleotide of AGCTU?
hypoxanthine? |
nucleoside = -oscine, nucleotide = -ylate
inosine, inosylate |
|
|
which has 2'-OH, which as 2'-H?
|
2'-OH = ribose, 2'-H = deoxyribose
|
|
|
glycosidic bond?
|
base + sugar
|
|
|
hydrophilic genetic components? Hydrophobic?
|
sugar + phosphate = hydrophilic
base = hydrophobic |
|
|
2 H bonds?
|
AT/AU
|
|
|
3 H bonds?
|
CG
|
|
|
double helix dimensions
|
3.4A (b/w bases), 34A (per turn of helix = 10 base pairs), 20A (diameter)
|
|
|
ssDNA vs dsDNA?
|
ssDNA (denatured) has a higher absorbance at 260 nm
|
|
|
melting temperature of DNA?
|
highly cooperative, increases with more CG (H bonds)
|
|
|
heterochromatin
|
histone < nucleosome < solenoid
|
|
|
histones
|
small basic (arginine/lysine rich) proteins
|
|
|
difference b/w uracil and thymine?
|
methyl group
|
|
|
RNA chains
|
single stranded but often display intraband base pairing
|
|
|
effect of alkali on DNA/RNA
|
DNA stays intact, RNA reduces to nucleotides (via nucleophilic attack by O- on phosphodiester bond)
|
|
|
components of mRNA
|
5'cap, start codon, stop codon, poly (A) tail… leader-->(coding) region-->trailer
|
|
|
prokaryote DNA polymerases
|
DNA Pol I = filling of gap after removal of RNA primer, DNA repair (3' to 5' exonuclease (proofreading) activity)
DNA Pol II = DNA Repair DNA Pol III = Replication of leading strand |
|
|
what fixed point does DNA start replication?
|
origin of replication, bi-direction in prokaryotes, multiple in eukaryotes
|
|
|
primase
|
synthesizes RNA primers
|
|
|
RNA primer
|
rest of DNA replication elongates from these
|
|
|
what removes RNA primers?
|
DNA Pol I
|
|
|
DNA ligase
|
restores phosphodiester bond
|
|
|
okazaki fragments
|
RNA primer -> DNA Pol III -> DNA ligase
|
|
|
helicase
|
unwinds DNA strand
|
|
|
SSB
|
binds to strand b/w helicase and DNA pol III
relieves tension? |
|
|
topoisomerase
|
relieves the strain induced by unwinding
|
|
|
beta-subunit of DNA Pol III
|
encircles DNA to enhance processivity
|
|
|
primosome
|
synthesizes RNA primers for okazaki strands
moves with replication fork |
|
|
eukaryote DNA polymerases
|
DNA Pol delta, alpha, beta, sigma, gamma
|
|
|
DNA Pol delta
|
Major Replicative Enzyme -Leading Strand
Has 3’ -> 5’ Proofreading Exonuclease Highly processive enzyme |
|
|
DNA Pol alpha
|
Polymerizing Activity works with Primase Activity
Lagging Strand Synthesis in complex with DNA Pol delta |
|
|
DNA Pol beta/sigma
|
DNA Repair
|
|
|
DNA Pol gamma
|
mitochondrial DNA synthesis
|
|
|
telomeres
|
(AGGGTT)n, n > 100
buffer at end of chromosome so when RNA primer is removed it doesn't take out genes, just repetitive sequence |
|
|
tautomers
|
bases go from normal state to less probable form leads to non-Watson/Crick base-pairing
hard for DNA repair |
|
|
spontaneous DNA damage
|
deamination (C -> U), depurination
fixed by replication |
|
|
chemical mutations
|
replaces purines/pyrimidines
|
|
|
physical mutations
|
dimers (T/T) caused by UV light, produces kink in helix
|
|
|
DNA repair accuracy
|
pushes back DNA mistakes form 10E-6 to 10E-10
recognizes daughter strand b/c parent strand is older, methylated in prokaryotes |
|
|
MutS/MutL and MutH
|
complex which binds parent/daughter strand, MutH comes in and cleaves unmethylated (daughter) strand
|
|
|
DNA glycosylases
|
recognize and remove abnormal bases, alkylated bases, pyrimidine dimers
|
|
|
AP endonuclease
|
cuts at apurinic or apyrimidic sites (AP) -> replaces with DNA Pol I -> nick sealed by ligase
|
|
|
why does DNA not have Uracil?
|
If DNA Normally had Uracil, Repair System Could not Distinguish Uracil that arose from Deamination of Cytosine from a Correct Uracil
No Mechanism to Guard Against GC -> AT Transitions that Result from Deamination of Cytosine to Uracil |
|
|
5-MeCyt
|
results in TG pairings, removed by GT mismatch glycosylase
5-MeCyt responsible for 1/3 of germline mutations |
|
|
nucleotide excision repair
|
endonuclease -> DNA Pol I -> DNA ligase
|
|
|
genetic defects in nucleotide excision repair
|
xeroderma pigmentosa, cockayne's syndrome, trichothiodystrophy
|
|
|
do both strands participate in transcription?
|
no, coding and template strands
coding strand 5'-3' = mRNA sequence (with Uracil) |
|
|
gene
|
consists of regions that are transcribed as well as regulatory sequences
|
|
|
promoter
|
initiation of transcription
|
|
|
template strand
|
non-coding strand
|
|
|
where is RNA synthesized in eukaryotes?
|
in the nucleus - mRNA by RNA Pol II, tRNA by RNA Pol II
in the nucleolus - rRNA by RNA Pol I |
|
|
prokaryotic promoters
|
TATAAT box (-10ish), TTGACA (-35)
|
|
|
sigma subunit
|
part of RNA Polymerase, recognize promoter and binds then leaves the larger unit
different sigma subunits confer recognition of different promoters influenced by environmental conditions |
|
|
transcription termination
|
hairpins (U-rich regions follow hairpin)
rho-dependent = ATPase binds nascent RNA chained and pulls it away from RNA polymerase |
|
|
differences b/w replication and transcription
|
Primer-Dependence
Proof-reading Activity of Polymerizing Enzyme NTPs vs. dNTPS Uracil vs. Thymine Conservative vs. Semi-conservative |
|
|
products of eukaryotic RNA polymerases
|
Pol I = rRNAs, Pol II = mRNAs, Pol III = tRNAs, short RNAs
|
|
|
eukaryotic promoters
|
TATA box (-25), GC-rich region (-40 to -80), CAAT box (-80 to -110)
|
|
|
TFII
|
transcription factors (help bind Pol II to DNA)
|
|
|
TBP
|
TATA Binding Protein (like sigma for RNA Polymerase)
|
|
|
TFIIH
|
recruits DNA repair proteins
|
|
|
transcription apparatus
|
TBP, TFIID, TFII(A/B), TFII(E/F/H), coactivators?
|
|
|
trans-acting factors
|
like DNA Pol II
|
|
|
cis-acting site
|
upstream of the gene, where trans-acting factors bind
|
|
|
prokaryote mRNAs are functional as transcribed
|
like polycistronic mRNA
|
|
|
tRNA cloverleaf
|
D (loop), anticodon (loop), variable loop, T/pseudouridine (loop), 5'CCA3' amino acid attachment site
|
|
|
nucleotidly transferase
|
|
|
|
45S Primary Transcript
|
Pol I (in nucleolus) produces 18S, 5.8S, and 28S -> organized (in nucleus) as 60s and transported out of nucleus
|
|
|
5S
|
no processing, produced by Pol III, nucleus
|
|
|
processing eukaryotic tRNAs
|
remove introns, ligase P-bonds, bases modified postranscriptionally
-primary transcripts fold prior to processing |
|
|
snRNP
|
roles in splicesome
bind 5' splice junction, bind branch site |
|
|
spliceosome
|
50-60S, contains snRNPs
|
|
|
intro functions
|
alternative splicing, exon shuffling
|
|
|
splice site mutation
|
beta-globin gene example
|
|
|
where does protein synthesis occur (in eukaryotes?)
|
cytosol
|
|
|
how many codons code for amino acids
|
61 + 3 stop codons
|
|
|
degenerate
|
many amino acids are specified by more than one codon (nucleotide triplet)
61 > 3 stop codons helps decrease mutations (as opposed to 20 aa codons and 44 stop codons) |
|
|
unambiguous
|
no codon specifies more than one amino acid
|
|
|
synonyms
|
most differ in only 3rd position
change in 1st position tend to specify similar aa's |
|
|
pyr in 2nd position
|
hydrophobic
|
|
|
pu in 2nd position
|
polar
|
|
|
wobble base
|
3' end of codon, 5' end of tRNA
|
|
|
inosine (I)
|
frequently found in the 5'position of the anticodon
contains hypoxanthine base pairs with U, C, A |
|
|
wobble hypothesis
|
"The Third Base of Codons Often Pairs Loosely with Corresponding Base of Anticodon - “Wobble Position”
Not all Base-Pairs are Allowed in the Wobble Position |
|
|
in wobble position, U pairs with:
|
A or G
|
|
|
in wobble position, G pairs with:
|
C or U
|
|
|
conservative missense mutation
|
encodes a similar amino acid
|
|
|
nonconservative missense mutation
|
encodes a non-similar amino acid
|
|
|
nonsense mutation
|
changes codon to a stop codon
|
|
|
read through mutations
|
changes a stop codon to an amino acid codon
|
|
|
aminoacyl-tRNA synthetase
|
enzymes that join aa's to their cognate tRNAs
20 synthetases, one for each aa specific for one amino acid and all of the tRNAs that carry |
|
|
purpose of tRNA?
|
make oxygen anion (on amino acid) a nucleophile by attaching it to tRNA (poor of LG)
tRNA activates aa for peptide bond formation |
|
|
aa-tRNA synthetase "charging rxn"
|
1. aa is activated via formation of an 5'-AMINOACYL ADENYLATE intermediate on the enzyme
2. the activated amino acid is transferred to the tRNA |
|
|
5'-aminoacyl adenylate
|
intermediate in aa-TRNA synthetase
|
|
|
ester linkage (tRNA)
|
activates the amino acid and joins it to the tRNA
|
|
|
hydrolytic "proofreading activity"
|
tRNA's ability to identify correct amino acid
|
|
|
met-tRNA(met) vs (fmet)
|
associates with AUG start codon
formylation of methionine (special property unique to initiator tRNA) fmet is eukaryote |
|
|
5'-UTR
|
5'-untranslated region (before AUG)
|
|
|
P-site
|
initial tRNA (met-tRNA) is bound, site of peptide bond formation
|
|
|
IF1
|
small ribosomal unit (translation) - prevents tRNA binding to the A site
|
|
|
IF3
|
prevents association of the large subunit
|
|
|
16S rRNA
|
associates with Shine-Delgarno sequence -> mRNA binds with AUG over P site of small ribosomal unit
|
|
|
IF2
|
met-tRNA associates with IF2 + GTP to bind to AUG site
|
|
|
what happens after initiating tRNA binds AUG?
|
large ribosomal unit (50S) joins small ribosomal unit
IF1, IF2, IF3, GDP, Pi leave |
|
|
eIF
|
eukaryotic initiation factor
|
|
|
CBP
|
cap binding protein complex (eukaryotic initiation)
|
|
|
EF-Tu/GTP
|
incoming aa-tRNA is bound
complex then binds A site |
|
|
peptidyltransferase
|
23S rRNA catalyzes peptide bond formation
|
|
|
A-site
|
dipeptidyl-tRNA
|
|
|
P-site
|
uncharged (deacylated) tRNA (during elongation)
|
|
|
translocation
|
moves ribosome to next codon while keeping tRNA's in A and P attached (shifts)
requires EF-G (translocase) and GTP |
|
|
EF-G (translocase)
|
requirement for translocation
|
|
|
eukaryotic elongation factors
|
EF-Tu, eEF-1(alpha)) = binds aa-tRNA site
EF-TS, eEF-1(Beta/gamma), GEF or GTP/GDP = exchange protein EF-G, eEF2 = translocase |
|
|
what has high affinity for A-site?
|
EF-Tu/GTP
|
|
|
what has low affinity for A-site
|
EF-Tu/GDP = hydrolyzed EF-Tu/GTP
|
|
|
what must happen before peptide bond forms?
|
EF-Tu released (hydrolyze EF-Tu/GTP)
|
|
|
GEF
|
EF-t's
Guanine Nucleotide Exchange Factor regenerates EF-Tu/GTP releases GDP |
|
|
release factors
|
enters A-site at AUG and terminates elongation
|
|
|
housekeeping genes
|
expressed in most cells most of the time
|
|
|
regulated genes
|
spatial
-tissue or cell-type specific temporal -specific stage of development |
|
|
constitutive expression
|
unregulated gene is expressed at the same level all the time
|
|
|
positive regulation of expression
|
inducible - gene expression increased
vs repressible |
|
|
polycistronic mRNA
|
contain their own SD sequence, start and stop codons
|
|
|
transcription factors
|
bind to operator
repressor (negative) or activators (positive) |
|
|
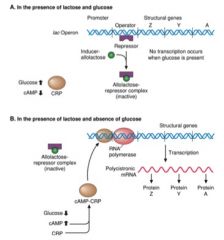
lac operon
|
repressor is only bound when lactose is missing
cAMP levels increase when glucose drops activator binds when cAMP is present |
|
|
CAP
|
cAMP-binding protein (= CRP)
|
|
|
CRP
|
catabolite repressor protein (= CAP)
|
|
|
catabolite repression
|
Glucose is the Preferred Carbon Source
Many Operons Involved in Sugar Catabolism (eg., lactose, galactose, arabinose) are not turned on even in the presence of the sugar if glucose is also available These Operons are Positively Regulated by the CAP (CRP) Protein The Signal for CAP binding is cAMP cAMP Levels Increase as Glucose Levels Drop |
|
|
DNA binding domains
|
60-90 aa's (small)
Domains Protrude From the Protein Surface to Interact with Bases in the Major Groove DNA Binding Sites are Often Inverted Repeats (Palindromes) and Regulatory Proteins are Often Dimers A Few Common Motifs |
|
|
helix-turn-helix
|
2 short a-helical segments separated by a beta turn
often dimers Two copies of the recognition helix are separated by one turn of DNA Helix dimers bind to palindromic site (like CRP) |
|
|
CRP site
|
palindromic (in promoter region)
|
|
|
leucine zipper
|
mediates both DNA binding and protein dimerization
|
|
|
features of eukaryotic transcription
|
Access to Promoters is Limited by the Structure of Chromatin
Positive Regulatory Mechanisms Predominate Regulatory Proteins are Larger and More Complex Transcription and Translation Occur in Different Sites |
|
|
HATs
|
histone acetyltransferases = helps prepare promoter regions for transcription
|
|
|
"hotspot" for mutation
|
Methylation of cytosine residues in GC boxes near promoters inhibits transcription.
|
|
|
how many cis-acting regulatory sequences on average in promoter?
|
six
|
|
|
transcriptional regulatory proteins
|
basal transcription factors, transactivators, cofactors
|
|
|
basal TFs
|
required by all RNA Pol II promoters
|
|
|
transactivators
|
1. Upstream Factors
Recognize Specific Elements in DNA (Upstream Activating Sequences, UASs) Activity of Factors is Not Regulated Ubiquitously expressed and act on any promoter that has the DNA Element 2. Inducible Factors Also recognize Specific Elements in DNA (Response Elements) synthesized or Activated at Specific Times or in Specific Places |
|
|
coactivators
|
Bridge the RNAP and Transactivators
|
|
|
TFII
|
transcription factor for RNA Pol II
|
|
|
response elements
|
bind the inducible factors under certain conditions when genes are expressed
make promoters subject to coordinate control |
|
|
cholesterol synthesis
|
Sterol Response Elements (SRE) bind inducible transactivators called SRE-Binding Proteins (SREBPs) to activate transcription
Sterol Levels Drop – Moves from the ER to the Golgi where proteases liberate the DNA Binding Domain, which Enters the Nucleus and Activates Transcription |
|
|
SREBPs
|
SRE-Binding Proteins (cholesterol synthesis):
1. ER membrane anchoring domains 2. helix-loop-helix DNA binding domain |
|
|
RNA editing
|
liver vs intestine
intestine does a point mutation on apoprotein B (ApoB) gene to get half the product |
|
|
heme-controlled kinase
|
When Heme Levels Drop, Kinase is Activated - PhosphorylateseIF2 and Shuts Down Translation of Globin mRNA
|
|
|
iron homeostasis
|
important for translational regulation, mRNA stability
Fe is Taken up by the Transferrin Receptor Excess Iron is Sequestered by Ferritin |
|
|
transferrin receptor
|
brings Fe into cell
|
|
|
ferritin
|
pushes out excess Fe
|
|
|
IRE-BP
|
binds IRE to prevent translation (low Fe), binds iron (high Fe)
when it is not binding, ferritin is produced to push out excess Fe |
|
|
basic tools/methods of recombinant DNA technology
|
gel electrophoresis, restriction enzyme digestion, DNA ligase, hybridization
|
|
|
restriction enzymes
|
DNA endonucleases made by prokaryotes
recognize specific short sequences of DNA and cleave both strands of DNA at or near the recognition site useful for carrying out digestion of DNA in a controlled and defined manner |
|
|
type II restriction enzymes nomenclature
|
Eco"R"V
Eco - refers to the bacterium for which the enzyme was isolated R - strain designation V - relates to the # of enzymes isolated from the organism |
|
|
type II restriction enzymes recognition site
|
palindromic
|
|
|
three types of cleavage ends
|
5'/3' protruding ends, blunt ends
|
|
|
restriction mapping
|
description of the locations of the restriction enzyme cleavage sites on a DNA molecule
useful for comparing with mutant DNA |
|
|
partial digest
|
part of restriction mapping, compare with complete digest to find correct gene locations
|
|
|
DNA ligase
|
joins two DNA termini by phosphodiester linkage
requirements: DNA should be double stranded, one terminus should have a 3' hydroxyl and the other should have a 5' phosphate |
|
|
opposite of denaturation?
|
hybridization
|
|
|
radio labeled DNA probe method?
|
hybridization
|
|
|
factors affecting nucleic acid hybridization
|
temperature, base composition of the hybridizing region, degree of sequence complementarity b/w the hybridizing molecules, composition of the hybridization rxn mix
|
|
|
how many H bonds b/w GC
|
three
|
|
|
how many H bonds b/w AT
|
two
|
|
|
hybridization positively affected by higher concentration of what?
|
GC
|
|
|
complementarity
|
greater complementarity increases efficient hybridization
|
|
|
what disrupts H bonds (weakens hybridization)
|
urea, formamide
|
|
|
what favors hybridization?
|
higher salt concentrations
|
|
|
stringency
|
hybridize at low stringency to hybridize, then increase stringency to find desired denatured product
|
|
|
southern analysis
|
enables the detection and analysis of a specific DNA sequence or gene in a DNA sample of extremely high sequence complexity
requires a probe containing the sequence of interest emlploys the principles of hybridization, electrophoresis, and restriction digestion |
|
|
RFLP
|
restriction fragment length polymorphism (RFLP) - southern analysis, DNA finger printing
|
|
|
southern analysis steps
|
gel electrophoresis -> denaturation -> transfer to membrane -> hybridization w/probe ->
|
|
|
VNTR
|
variable number of tandem repeats
repetitive sequences present in different loci in the human gene DNA finger printing |
|
|
PCR
|
polymerase chain reaction
Enables the amplification of a specific segment of DNA starting from extremely low amounts of DNA Requires very little sequence information regarding the sites flanking the segment to be amplified Allows human identity testing from extremely small samples (e.g., few drops of blood, cells obtained from the roots of few hairs etc.,) Allows detection of pathogenic bacteria and viruses from very small biological samples (e.g., tubercle bacilli) |
|
|
PCR steps
|
Mix dNTPs, template, primers and DNA polymerase
Heat the reaction mix to ~94°C to denature template Cool the reaction mix to a temperature at which the primers can hybridize to the template (~50 to 65°C) Heat the reaction mix to the temperature that is optimum for elongation of the primers by the polymerase Repeat 25 to 35 times (heat stable Taq polymerase used) |
|
|
PCR to diagnose Duchenne Muscular Dystrophy (DMD)
|
DMD is caused by deletions in specific exons of Dystrophin gene
results in PCR products of different sizes…DMD patients will produce smaller PCR product |
|
|
PCR with point mutations
|
PCR forward primer is allele specific, reverse primer is not allele specific
e.g. normal individual will produce PCR results with forward primer if it comes across a point mutation A-->G (cystic fibrosis), but not in normal individuals so, if there are PCR strands, there is a mutation. |
|
|
Single Strand Conformational Polymorphism (SSCP)
|
Folded structures (formed by intramolecular base pairing) adopted by the single stranded nucleic acid molecules are determined by their primary sequences
Single stranded DNA molecules that fold into different structures exhibit different mobilities during electrophoresis through a native gel |
|
|
Denaturing Gradient Gel Electrophoresis (DGGE)
|
Mobility of double stranded DNA in electrophoresis through a denaturing gradient gel can be affected by very small changes in its primary sequence
|
|
|
Heteroduplex analysis
|
Finding mismatches by mixing DNA from patient with normal DNA (mix-->denature-->renature-->analyze PCR products)
|
|
|
Sanger's method
|
most widely used DNA sequencing method
create a specific primer, mix with template, take largest product to determine the rest of the sequence (be careful of 5'-3') |
|
|
dideoxy NTP
|
terminates elongation
|
|
|
ddATP, ddGTP, ddCTP, ddTTP
|
|
|
|
basic tools/methods of recombinant DNA technology
|
gel electrophoresis, restriction enzyme digestion, DNA ligase, hybridization
|
|
|
restriction enzymes
|
DNA endonucleases made by prokaryotes
recognize specific short sequences of DNA and cleave both strands of DNA at or near the recognition site useful for carrying out digestion of DNA in a controlled and defined manner |
|
|
type II restriction enzymes nomenclature
|
Eco"R"V
Eco - refers to the bacterium for which the enzyme was isolated R - strain designation V - relates to the # of enzymes isolated from the organism |
|
|
type II restriction enzymes recognition site
|
palindromic
|
|
|
three types of cleavage ends
|
5'/3' protruding ends, blunt ends
|
|
|
restriction mapping
|
description of the locations of the restriction enzyme cleavage sites on a DNA molecule
useful for comparing with mutant DNA |
|
|
partial digest
|
part of restriction mapping, compare with complete digest to find correct gene locations
|
|
|
DNA ligase
|
joins two DNA termini by phosphodiester linkage
requirements: DNA should be double stranded, one terminus should have a 3' hydroxyl and the other should have a 5' phosphate |
|
|
opposite of denaturation?
|
hybridization
|
|
|
radio labeled DNA probe method?
|
hybridization
|
|
|
factors affecting nucleic acid hybridization
|
temperature, base composition of the hybridizing region, degree of sequence complementarity b/w the hybridizing molecules, composition of the hybridization rxn mix
|
|
|
how many H bonds b/w GC
|
three
|
|
|
how many H bonds b/w AT
|
two
|
|
|
hybridization positively affected by higher concentration of what?
|
GC
|
|
|
complementarity
|
greater complementarity increases efficient hybridization
|
|
|
what disrupts H bonds (weakens hybridization)
|
urea, formamide
|
|
|
what favors hybridization?
|
higher salt concentrations
|
|
|
stringency
|
hybridize at low stringency to hybridize, then increase stringency to find desired denatured product
|
|
|
southern analysis
|
enables the detection and analysis of a specific DNA sequence or gene in a DNA sample of extremely high sequence complexity
requires a probe containing the sequence of interest emlploys the principles of hybridization, electrophoresis, and restriction digestion |
|
|
RFLP
|
restriction fragment length polymorphism (RFLP) - southern analysis, DNA finger printing
|
|
|
southern analysis steps
|
gel electrophoresis -> denaturation -> transfer to membrane -> hybridization w/probe ->
|
|
|
VNTR
|
variable number of tandem repeats
repetitive sequences present in different loci in the human gene DNA finger printing |
|
|
PCR
|
polymerase chain reaction
Enables the amplification of a specific segment of DNA starting from extremely low amounts of DNA Requires very little sequence information regarding the sites flanking the segment to be amplified Allows human identity testing from extremely small samples (e.g., few drops of blood, cells obtained from the roots of few hairs etc.,) Allows detection of pathogenic bacteria and viruses from very small biological samples (e.g., tubercle bacilli) |
|
|
PCR steps
|
Mix dNTPs, template, primers and DNA polymerase
Heat the reaction mix to ~94°C to denature template Cool the reaction mix to a temperature at which the primers can hybridize to the template (~50 to 65°C) Heat the reaction mix to the temperature that is optimum for elongation of the primers by the polymerase Repeat 25 to 35 times (heat stable Taq polymerase used) |
|
|
PCR to diagnose Duchenne Muscular Dystrophy (DMD)
|
DMD is caused by deletions in specific exons of Dystrophin gene
results in PCR products of different sizes…DMD patients will produce smaller PCR product |
|
|
PCR with point mutations
|
PCR forward primer is allele specific, reverse primer is not allele specific
e.g. normal individual will produce PCR results with forward primer if it comes across a point mutation A-->G (cystic fibrosis), but not in normal individuals so, if there are PCR strands, there is a mutation. |
|
|
Single Strand Conformational Polymorphism (SSCP)
|
Folded structures (formed by intramolecular base pairing) adopted by the single stranded nucleic acid molecules are determined by their primary sequences
Single stranded DNA molecules that fold into different structures exhibit different mobilities during electrophoresis through a native gel |
|
|
Denaturing Gradient Gel Electrophoresis (DGGE)
|
Mobility of double stranded DNA in electrophoresis through a denaturing gradient gel can be affected by very small changes in its primary sequence
|
|
|
Heteroduplex analysis
|
Finding mismatches by mixing DNA from patient with normal DNA (mix-->denature-->renature-->analyze PCR products)
|
|
|
Sanger's method
|
most widely used DNA sequencing method
create a specific primer, mix with template, take largest product to determine the rest of the sequence (be careful of 5'-3') |
|
|
dideoxy NTP
|
terminates elongation
|
|
|
ddATP, ddGTP, ddCTP, ddTTP
|
|
|
|
oligonucleotides
|
short strands (<80 nucleotides)
Primers for PCR and DNA sequencing Probes for Southern and Northern analyses ‘Linkers’ that add restriction sites to DNA molecules |
|
|
Linker DNA
|
Linkers help to make the DNA ends compatible for ligation
|
|
|
gene cloning purpose
|
recombinant DNA methods -> isolation -> characterization
|
|
|
gene cloning steps
|
1. fragmentation of DNA
2. genomic DNA fragments have to be inserted into a cloning vector to construct genomic library 3. transformation - introduction of plasmid DNA into bacterial host cells (cells that do not take up plasmid do not grow on ampicillin containing medium) 4. recombinant vectors separated from empty vectors - one way to do this is through beta-galactosidase (blue/white dots on plate, white = foreign gene inserted) |
|
|
bacteriophage lambda
|
efficient cloning vector
recombinant phage infects bacteria w/target gene |
|
|
cDNA library exception
|
cDNA library represents only those genes expressed in the tissue from which they it was made
|
|
|
sequences needed for efficient expression of genes in bacteria
|
Promoter and transcription start site
Sequences coding for ribosome binding site on mRNA Transcription termination site - These sequence elements are absent in cDNA - Expression vectors are plasmid vectors that provide such sequences and thereby allow the expression of the cDNA inserted in them. |
|
|
cDNA library screening
|
typically use hybridization and a labeled DNA probe
|
|
|
problems w/ eukaryotic proteins purified from bacteria
|
improper folding, no post-translational modifications (inactive), toxic bacterial proteins copurify w/protein
solution: yeast cells |
|
|
ways to introduce DNA into mammalian cells
|
transfection, microinjection, liposome-mediated delivery, viral vectors
|
|
|
advantage of using retroviral vectors in gene therapy
|
uses reverse transcriptase and integrase
DNA copy of viral genome is integrated into host cell's chromosome by replacing viral genes with gene of interest, viral gene cannot be replicated once successfully placed in host cell = recombinant (defective) retroviral DNA aka retroviral vector DNA note: helper virus DNA |
|
|
helper virus DNA
|
Helper virus DNAs cannot direct the packaging of their own RNA genomes but allow the packaging of vector virus RNA genome
|
|
|
Formation of replication competent virus particles is prevented in the procedure because:
|
Vector virus particle can enter cells but cannot multiply.
Helper virus RNA cannot be packaged since it lacks packaging sequence (Y). Each helper virus genome codes only for a subset of viral genes. Therefore, packaging of vector viral RNA can occur only in the presence of both helper virus genomes. |
|
|
Severe combined immunodeficiency-X1 (SCID-X1)
|
caused by defect in cytokine receptor g-c subunit gene
Lymphoid progenitor cell in SCID-X1 patients with defective gene for gcsubunit makes functionally impaired receptor |
|
|
Strategies for treatment of SCID:
|
Transplant bone marrow from a HLA-identical sibling to the patient
If a HLA-dentical sibling is not available, alternate treatment procedures include GENE THERAPY (Treated cells compete out defective cells due their growth advantage and give lasting relief) |
|
|
ex vivo vs in vivo
|
types of gene transfer
|
|
|
"knock out" mice
|
Mice carrying gene disruption (`knock out’ mice) serve as model systems for studying diseases and for testing treatment procedures for the diseases
|
|
|
gene disruption
|
Gene disruption makes use of the process of homologous recombination
|
|
|
DNA microarrays
|
DNA microarrays are useful in determining the mRNA expression profiles
mRNA expression profile is characteristic of a given cell type at a given physiological state Differences between the mRNA expression profiles of normal and diseased cells often provide useful insight into the basis of the disease |
|
|
DNA microarrays (steps)
|
Isolate mRNAs from normal and diseased cells.
Copy mRNAs into cDNA using reverse transcriptase and fluorescently labeled dNTPs. Use different colored fluorescent labels for the two mRNA samples Mix and Hybridize the labeled cDNAs to microarrays Wash away unhybridized probe and quantitate the fluorescence at each spot on the array |
|
|
Xeroderma Pigmentosum
|
mostly due to environmental influence (UV light)
freckling or burns freckling under premature aging of skin clouding of the eye/loss of eyelashes equal distribution m/f 97% of basal & squamous cells carcinoma occur on face, head, or neck (65% of melanomas occur in these regions) cancers occur <10 years pol eta defect UV is absorbed by cornea and lens (leads to eye cancers) neurological abnormalities due to nerve cell death (from UV light) SKIN CANCER |
|
|
Cockayne Syndrome
|
no skin pigmentation/cancer, no environment effect
mostly due by developmental defect calcification in basal ganglia NEUROLOGICAL DEFECTS |
|
|
Trichothiodystrophy
|
mostly due by developmental defect
calcification in basal ganglia rare autosomal recessive disorder short, brittle hair (tricho-) with low sulfur (-thio-) content broad spectrum of clinical phenotypes ichthyosis - fish scale, dry skin, photosensitive to sunlight glucodystrophy SHORT BRITTLE HAIR, DRY SKIN |
|
|
cyclobutane pyrmidine dimer (CPD)
|
75% of lesions in DNA
|
|
|
[6-4] PP
|
25% of lesions in DNA
|
|
|
XP-D (ERCC2)
|
10 different clinical disorders (grand daddy defect, encompasses all clinical disorders covered today)
|
|
|
XPG GENE
|
framehshift mutation, leads to nonsense (died at 6 y.o, neuro defects) or missense mutations (xeroderma pigmentosum, no neuro effects)
|
|
|
explain lac operon in the presence of glucose and lactose
|
|

