![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
57 Cards in this Set
- Front
- Back
|
Introns
|
are only found in eukaryotic genes.
|
|
|
The central dogma of biology
|
DNA -> RNA -> protein
|
|
|
HDAC
|
stands for histone deacetylase and is responsible for the deacetylation of histones.
|
|
|
HAT
|
Histone acetyltransferase and is responsible for acetylating histones.
|
|
|
Regulation of chromatin structure
|
is achieved through regulating the enzymatic activity of HDAC and HAT.
|
|
|
Transcription Factors
|
control gene expression by regulating or inhibiting gene transcription.
|
|
|
RNA processing
|
consist of capping, splicing, polyadenylation, and RNA editing.
|
|
|
What are the four elements found within an intron?
|
a 5', 3' splice site, branch point, and a polypyrimidine tract.
|
|
|
Can splicing be regulated?
|
Yes
|
|
|
How is splicing regulated?
|
Through the activity of SR proteins and hnRNP proteins.
|
|
|
(True or False) Alternative splicing is a way to increase protein diversity.
|
True
|
|
|
(True or False) When iron levels are low, ferritin mRNA is less stable.
|
True
|
|
|
(True or False) When iron levels are high, Ferritin mRNA has a longer half-life.
|
True
|
|
|
(True or False) When iron levels are low, TfR (transferrin receptor) mRNA has a short-half life.
|
False
|
|
|
(True or False) When iron levels are high, TfR mRNA is less stable.
|
True
|
|
|
(True or False) Nonsense mediated decay is a pathway for regulating RNA degradation.
|
True
|
|
|
Prader-Willi Syndrome
|
results from a paternal deletion of chromosome 15 q11-q13.
|
|
|
Angleman syndrome
|
results from a maternal deletion of chromosome 15 q11-q13.
|
|
|
DNA methylation commonly occurs in:
|
GC rich regions of the promoter.
|
|
|
(True or False) DNA methylation is method of turning off gene expression.
|
True.
|
|
|
A prokaryotic operon consist of:
|
a promoter and structural genes.
|
|
|
(True or False) Transcription and Translation in Prokaryotic organisms are coupled?
|
True
|
|
|
(True or False) In prokaryotes, a repressor is responsible for inhibiting gene expression.
|
True
|
|
|
HDAC will inactivate or activate gene expression.
|
Inactivate
|
|
|
HAT will inactivate or activate gene expression.
|
Activate
|
|
|
What is the amino acid that is specifically acetylated by HAT.
|
Lysine
|
|
|
The conversion of C to U in Apoprotein produces:
|
Apo B48
|
|
|
A message that is not properly spliced will most likely be:
|
degraded by the NMD pathway.
|
|
|
Is prokaryotic DNA complexed w/ histones?
|
No; prokaryotes do not have a nuclear envelope separating genes from cytoplasmic contents. Prokaryotic gene transcripts do not contain introns.
|
|
|
What differentiates mRNA processing in prokaryotes from eukaryotes?
|
As prokaryotic mRNA is being synthesized, ribosomes bind and begin to produce proteins so that transcription and translation occur simultaneously... thus regulation at the level of initiation is sufficient to regulate the level of proteins within the cell.
|
|
|
What is the fnx of operons in the bacterial genome?
|
Genes that encode proteins are called 'structural genes'; structural genes for proteins involved in related functions are grouped sequentially into units called operons.
|
|
|
"all or none" operon fnx
|
The genes in operons are expressed coordinately; they are either all turned on or all turned off. When an operon is expressed, all of its genes are transcribed. A single polycistronic mRNA is produced that codes for all of the protein of an operon.
|
|
|
prokaryotic gene structure
|
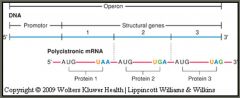
|
|
|
promoter
|
A specific DNA sequence to which RNA polymerase binds. Regulates transcription of genes in an operon.
-Located in the operon 5' end, upstream from the structural genes. |
|
|
repressors
|
-Regulatory proteins that prevent the binding of RNA polymerase to the promoter, preventing transcription.
-Principal means of regulating gene transcription in bacteria. |
|
|
operator
|
-Site of repressor binding to promtoers
-Located upstream of the transcription start point at the 3' end |
|
|
inducer
|
-inactivates repressors- "induction"
-stimulates expression of operon by binding repressor, preventing it from binding to the operator -Ex: induction of lac operon by lactose; when milk sugar lactose is available, enzymes for lactose maetabolism are produced which are encoded by the lac operon. A metabolite of lactose acts as an inducer binding to the repressor and inactivating it, allowing RNA polymerase to bind to the promoter so that transcription of the structural genes for the lac operon can be transcribed. |
|
|
What's the difference between heterochromatin and euchromatin?
|
-heterochromatin: condensed, inactive
-euchromatin: unwound, produces mRNA |
|
|
What does HDAC do?
|
histone deacetylase does as its name suggests; deacetylates lysine residues making histone cores positive allowing DNA to wrap around them
|
|
|
What does methylating DNA do?
|
Cytosine residues in DNA can be methylated in GC rich regions, often near the promoter. Methylated genes are less readily transcribed.
|
|
|
gene rearrangement
|
allows for DNA segments to move to other locations in the genome; for antibodies, this allows for tons of recombinational possibilities allowing for a much more diverse B cell population
|
|
|
What 3 processes are involved in processing of the primary transcript?
|
-Capping the 5' end
-Removal of introns -polyadenylation: addition of poly(A) tail to the 3' end |
|
|
alternative splicing
|
splicing exons together from the same gene in different orders to make different proteins; 75% of our genome is alternatively spliced
|
|
|
What does capping do?
|
-prevents 5'-3' exonucleases from degrading mRNA; controls mRNA expression
-allows mRNA to be transported from nucleus |
|
|
What does a poly(A) tail do?
|
-serves as a 3' linker which decreases 3'-5'degradation
-stabilizes mRNA -poly(A) tail length directly correlates with the 1/2 life of the mRNA |
|
|
What 2 parts of introns are invariable?
|
-5' GU & 3' AG
-branch point adensoine |
|
|
in the human genome, which is longer, exons or introns?
|
introns, even though exons are what actually encode protein
|
|
|
ESE & ESS
|
exon splicing enhancers & suppressors
|
|
|
SR proteins & hnRNPs
|
activate & inhibit splicing, respectively
|
|
|
nonsense mediated decay
|
failure to remove introns results in degradation of mRNA
|
|
|
RNA editing
|
addition of stop codon to an mRNA transcript resulting in different translation products, e.g. ApoB48 & ApoB100: stop codon UAA added to ApoB protein resulting in protein length 48% of the other.
|
|
|
ferritin
|
(intracellular) stores iron; ferritin mRNA translation increases w/ increasing levels of iron so that cells can store more iron
|
|
|
transferrin
|
(extracellular) transports iron; transferin receptor mRNA degradation increases w/ increasing levels of iron because less iron needs to be taken in by cells
|
|
|
iron response element binding protein (IRE-BP)
|
-low levels of iron: IRE-BP binds and stabilizes transferrin receptor mRNA 3' site & destabilizes ferritin receptor mRNA 5' site
-high iron levels: IRE-BP binds iron decreasing its affinity for mRNA; dissociation destabilizes transerrin receptor mRNA & stabilizes ferritin receptor mRNA |
|
|
Schmid metaphyseal chondrodysplasia
|
autosomal dominant skeletal dysplasia characterized by short stature, coxa vara, genu varum and a wide irregular growth plate. SMCD results from heterozygous mutations in the gene for collagen X, a short-chain collagen whose expression is largely restricted to the hypertrophic chondrocytes of growth plate cartilage.
|
|
|
Prader-Willi syndrome (2)
|
chromosome 15 PATERNAL genes 1,2 & 3 are deleted and maternal gene 2 is methylated resulting in expression of maternal genes 1 & 3 only
|
|
|
Angelman syndrome (2)
|
chromosome 15 MATERNAL genes 1,2 & 3 are deleted and paternal gene 1 is methylated resulting in expression of paternal genes 2 & 3 only
|

