![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
18 Cards in this Set
- Front
- Back
|
rRNAs are members of the ______ sequence class.
|
intermediate
|
|
|
Oscar Miller feathers
|
multiple transcripts from each gene are produced simultaneously... they like the barbs of a feather and Oscar Miller discovered 'em, so there you go.
|
|
|
The _____ is the region of a nucleus where ribosomes are produced.
|
nucleolus; not membrane bound
-region where multiple rRNA transcripts are expressed from multiple genes, with protein binding each transcript. With so many ribosomes “pouring out,” the region looks dark under a light microscope. |
|
|
Each gene encodes ___ rRNAs. They are all transcribed by Pol __ as one large RNA molecule, known as Pre-rRNA. The intervening sequences are then cleaved away, leaving the three rRNAs; 28S, 18S, and 5.8S.
|
3, Pol I
|
|
|
What 3 rRNAs are left after intervening sequences are cleaved away?
|
-28S
-18S -5.8S |
|
|
A forth rRNA is transcribed from separate _____ genes. These are transcribed by Pol ___, the polymerase that produces tRNAs.
|
5S rRNA, Pol III
|
|
|
ribonucleoprotieins
|
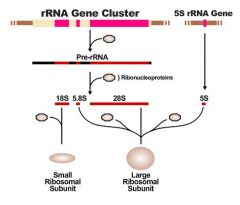
proteins that assemble around the rRNA to form the large and small subunits of ribosomes. Therefore, ribosomes are composed of RNA and protein.
|
|
|
What makes up the large ribosomal subunit? What about the small ribosomal subunit?
|
-large: 5S, 5.8S, 28S + ribonucleoproteins
-small- 18S + ribonucleoproteins |
|
|
Each rRNA gene is polycistronic,meaning is encodes multiple _____.
|
rRNA transcripts
|
|
|
Polycistronic rRNAs are transcried as intact units call _____.
|
pre-rRNAs
|
|
|
tRNA structure
|
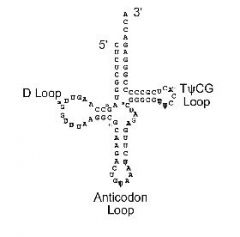
The primary transcripts of tRNAs undergo extensive post transcriptional modification. This includes intron splicing, nucleotide addition, methylation, and modifications to produce unusual nucleotides such as dihydrouridine (D), pseudouridine (ψ) and ribothymidine (T).
|
|
|
folding
|
Internal annealing folds tRNAs into “clover leaves.” The D and TψCG loops also fold back on each other to generate L shaped structures
|
|
|
Anticodon
|
A nucleotide triplet at the apex of the tRNA, which is complimentary to a specific codon of mRNA. In the ribosome, an anticodon binds a codon. This is how codons specify amino acids
|
|
|
Unique properties of RNA polymerase I and Pol III promoters
|
Unlike Pol II, promoters for rRNA and tRNA lack TATA boxes. Surprisingly, TBP is still a component of their initiation complexes.
|
|
|
Pol III
|
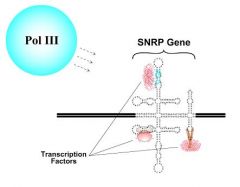
Pol III promoters are within their genes. Moreover, their DNA strands fold back on themselves to form internal base pairs. Note, the small RNAs transcribed by Pol III are characterized by internal annealing. Therefore, Pol III recognizes the same tertiary structure in the DNA that exists in the RNA.
|
|
|
nuclear pore complexes
|
mRNA, tRNA and ribosomes are all produced in the nucleus, but translation occurs in the cytoplasm, so all three components must be transported out. Other factors, such as DNA, transcription factors, and RNA polymerase must be imported into the nucleus. This occurs through channels embedded in the nuclear envelope, known as nuclear pore complexes. These are huge structures, 16 times larger than ribosomes, composed of 30 different proteins known as nucleoporins. The selective transport of factors into or out of the nucleus is mediated by proteins such as importins and exportins, which bind specific factors, and shuttle them through the nuclear pore complex.
|
|
|
tRNA charging
|
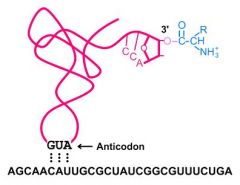
After tRNAs reach the cytoplasm, there is one more step before they are ready to be used for translation. They must be bound with their specific amino acids. This is often referred to as tRNA loading or charging. An enzyme called an aminoacyl-tRNA synthetase binds the 3' OH of the tRNA with the carboxyl group of its amino acid. The reaction requires an ATP for energy.
|
|
|
degeneracy of the genetic code
|
there are over 50 tRNAs, but there are only 20 amino acids. So a given amino acid can be bound to several tRNAs. However, each tRNA is only bound to one amino acid, and will only bind one codon. Multiple tRNAs for a given amino acid contributes to the degeneracy of the genetic code.
|

